
Capybara microvirus Cap1_SP_206
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.54
Get precalculated fractions of proteins
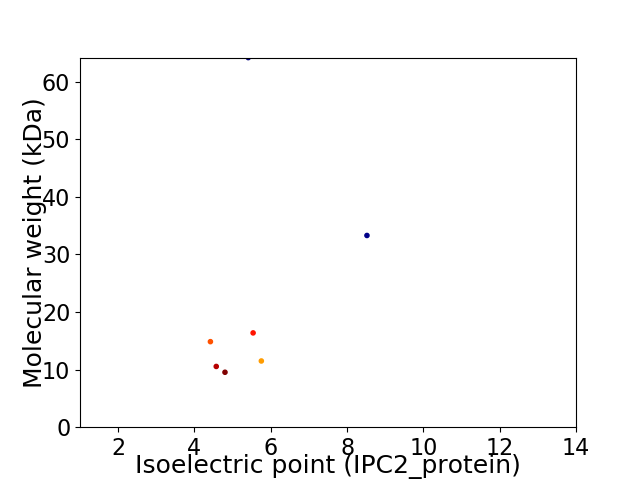
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5F1|A0A4P8W5F1_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_206 OX=2585409 PE=4 SV=1
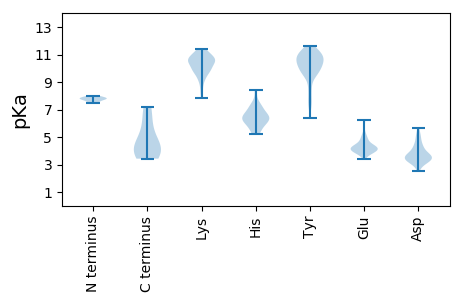
MM1 pKa = 7.77SDD3 pKa = 3.7SKK5 pKa = 11.16FDD7 pKa = 3.82PSKK10 pKa = 11.17DD11 pKa = 3.4FEE13 pKa = 6.26LDD15 pKa = 3.28DD16 pKa = 4.8GSIVHH21 pKa = 6.34YY22 pKa = 10.46GDD24 pKa = 3.25PRR26 pKa = 11.84YY27 pKa = 10.38IFDD30 pKa = 4.39IGWNDD35 pKa = 3.31EE36 pKa = 4.26SKK38 pKa = 11.22KK39 pKa = 10.92DD40 pKa = 3.56FLIIKK45 pKa = 8.34GAQPFNEE52 pKa = 4.2VMDD55 pKa = 3.74KK56 pKa = 10.84RR57 pKa = 11.84AIGTTLYY64 pKa = 10.83EE65 pKa = 4.58LIDD68 pKa = 4.0RR69 pKa = 11.84YY70 pKa = 11.06GGVDD74 pKa = 3.14EE75 pKa = 5.32CSAAFSEE82 pKa = 4.49GGVYY86 pKa = 10.54ADD88 pKa = 3.52VVGTPEE94 pKa = 3.83FGNNEE99 pKa = 3.78SYY101 pKa = 11.21AAVLHH106 pKa = 5.24QLKK109 pKa = 10.53DD110 pKa = 3.48QLSKK114 pKa = 11.39LEE116 pKa = 4.07EE117 pKa = 4.04ANKK120 pKa = 10.19QIEE123 pKa = 4.37KK124 pKa = 7.85TTEE127 pKa = 3.77KK128 pKa = 10.99KK129 pKa = 10.6EE130 pKa = 3.87IEE132 pKa = 4.14
MM1 pKa = 7.77SDD3 pKa = 3.7SKK5 pKa = 11.16FDD7 pKa = 3.82PSKK10 pKa = 11.17DD11 pKa = 3.4FEE13 pKa = 6.26LDD15 pKa = 3.28DD16 pKa = 4.8GSIVHH21 pKa = 6.34YY22 pKa = 10.46GDD24 pKa = 3.25PRR26 pKa = 11.84YY27 pKa = 10.38IFDD30 pKa = 4.39IGWNDD35 pKa = 3.31EE36 pKa = 4.26SKK38 pKa = 11.22KK39 pKa = 10.92DD40 pKa = 3.56FLIIKK45 pKa = 8.34GAQPFNEE52 pKa = 4.2VMDD55 pKa = 3.74KK56 pKa = 10.84RR57 pKa = 11.84AIGTTLYY64 pKa = 10.83EE65 pKa = 4.58LIDD68 pKa = 4.0RR69 pKa = 11.84YY70 pKa = 11.06GGVDD74 pKa = 3.14EE75 pKa = 5.32CSAAFSEE82 pKa = 4.49GGVYY86 pKa = 10.54ADD88 pKa = 3.52VVGTPEE94 pKa = 3.83FGNNEE99 pKa = 3.78SYY101 pKa = 11.21AAVLHH106 pKa = 5.24QLKK109 pKa = 10.53DD110 pKa = 3.48QLSKK114 pKa = 11.39LEE116 pKa = 4.07EE117 pKa = 4.04ANKK120 pKa = 10.19QIEE123 pKa = 4.37KK124 pKa = 7.85TTEE127 pKa = 3.77KK128 pKa = 10.99KK129 pKa = 10.6EE130 pKa = 3.87IEE132 pKa = 4.14
Molecular weight: 14.85 kDa
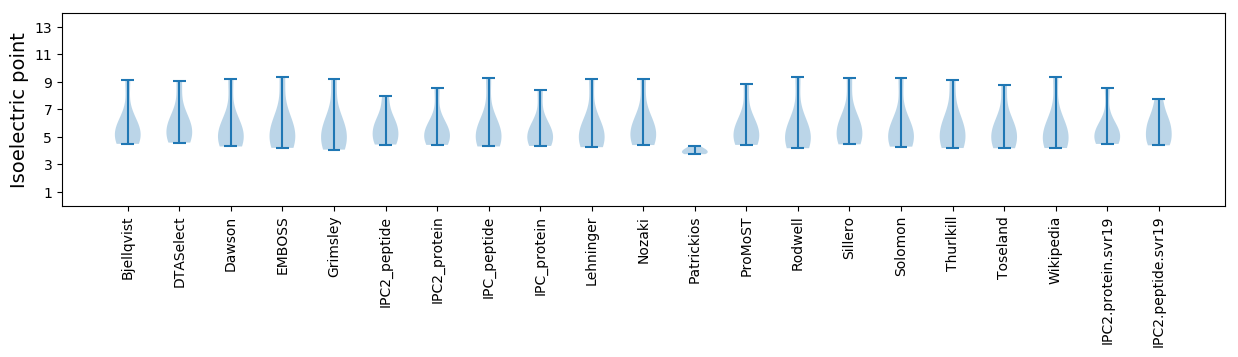
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVR0|A0A4V1FVR0_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_206 OX=2585409 PE=4 SV=1
MM1 pKa = 7.86CLRR4 pKa = 11.84PLHH7 pKa = 5.99YY8 pKa = 10.06FQSDD12 pKa = 2.76IMNEE16 pKa = 4.3STGKK20 pKa = 10.63YY21 pKa = 8.28YY22 pKa = 11.21GFVTGYY28 pKa = 7.49QTLVADD34 pKa = 4.07YY35 pKa = 11.16NDD37 pKa = 3.42FKK39 pKa = 11.43LRR41 pKa = 11.84DD42 pKa = 3.75IPCTLAHH49 pKa = 7.12RR50 pKa = 11.84ATPIKK55 pKa = 9.77VACGRR60 pKa = 11.84CCEE63 pKa = 4.27CLNAKK68 pKa = 9.61KK69 pKa = 10.07MSWVGRR75 pKa = 11.84AVAEE79 pKa = 4.46LEE81 pKa = 4.21TSKK84 pKa = 11.15YY85 pKa = 10.95GYY87 pKa = 10.02FLSITYY93 pKa = 10.75NNDD96 pKa = 2.55HH97 pKa = 6.57VLPKK101 pKa = 9.92PEE103 pKa = 3.97KK104 pKa = 10.51RR105 pKa = 11.84EE106 pKa = 3.49IQLFIKK112 pKa = 10.48RR113 pKa = 11.84LRR115 pKa = 11.84KK116 pKa = 9.38HH117 pKa = 5.85FKK119 pKa = 10.25LRR121 pKa = 11.84YY122 pKa = 7.31LTVGEE127 pKa = 4.85LGSLTDD133 pKa = 3.42RR134 pKa = 11.84AHH136 pKa = 5.85YY137 pKa = 10.78HH138 pKa = 6.02MILYY142 pKa = 10.39SNQPINDD149 pKa = 3.55LVYY152 pKa = 10.45YY153 pKa = 10.18SSSGSNILYY162 pKa = 9.56TSEE165 pKa = 6.02LIDD168 pKa = 3.75SCWQKK173 pKa = 11.3GSIKK177 pKa = 9.94IGRR180 pKa = 11.84AEE182 pKa = 4.16ASSIAYY188 pKa = 6.41TVGYY192 pKa = 9.7IVSKK196 pKa = 10.28EE197 pKa = 3.75KK198 pKa = 9.66KK199 pKa = 8.19TCFKK203 pKa = 10.72LQSQGLGFDD212 pKa = 3.45YY213 pKa = 10.64FKK215 pKa = 11.11NLNSSYY221 pKa = 11.04VLSTGRR227 pKa = 11.84GKK229 pKa = 10.27EE230 pKa = 3.85LYY232 pKa = 10.19VRR234 pKa = 11.84LPRR237 pKa = 11.84YY238 pKa = 9.69LKK240 pKa = 10.28EE241 pKa = 3.99KK242 pKa = 10.62YY243 pKa = 9.78GLKK246 pKa = 10.24SEE248 pKa = 4.21FDD250 pKa = 3.49SVKK253 pKa = 10.86AEE255 pKa = 4.0RR256 pKa = 11.84EE257 pKa = 4.07WKK259 pKa = 10.57NKK261 pKa = 9.21VFGSGLDD268 pKa = 3.48EE269 pKa = 4.07EE270 pKa = 5.4DD271 pKa = 3.54YY272 pKa = 11.45RR273 pKa = 11.84DD274 pKa = 3.72FKK276 pKa = 11.34QYY278 pKa = 11.04LSEE281 pKa = 4.37HH282 pKa = 6.37KK283 pKa = 10.67LIVHH287 pKa = 7.16
MM1 pKa = 7.86CLRR4 pKa = 11.84PLHH7 pKa = 5.99YY8 pKa = 10.06FQSDD12 pKa = 2.76IMNEE16 pKa = 4.3STGKK20 pKa = 10.63YY21 pKa = 8.28YY22 pKa = 11.21GFVTGYY28 pKa = 7.49QTLVADD34 pKa = 4.07YY35 pKa = 11.16NDD37 pKa = 3.42FKK39 pKa = 11.43LRR41 pKa = 11.84DD42 pKa = 3.75IPCTLAHH49 pKa = 7.12RR50 pKa = 11.84ATPIKK55 pKa = 9.77VACGRR60 pKa = 11.84CCEE63 pKa = 4.27CLNAKK68 pKa = 9.61KK69 pKa = 10.07MSWVGRR75 pKa = 11.84AVAEE79 pKa = 4.46LEE81 pKa = 4.21TSKK84 pKa = 11.15YY85 pKa = 10.95GYY87 pKa = 10.02FLSITYY93 pKa = 10.75NNDD96 pKa = 2.55HH97 pKa = 6.57VLPKK101 pKa = 9.92PEE103 pKa = 3.97KK104 pKa = 10.51RR105 pKa = 11.84EE106 pKa = 3.49IQLFIKK112 pKa = 10.48RR113 pKa = 11.84LRR115 pKa = 11.84KK116 pKa = 9.38HH117 pKa = 5.85FKK119 pKa = 10.25LRR121 pKa = 11.84YY122 pKa = 7.31LTVGEE127 pKa = 4.85LGSLTDD133 pKa = 3.42RR134 pKa = 11.84AHH136 pKa = 5.85YY137 pKa = 10.78HH138 pKa = 6.02MILYY142 pKa = 10.39SNQPINDD149 pKa = 3.55LVYY152 pKa = 10.45YY153 pKa = 10.18SSSGSNILYY162 pKa = 9.56TSEE165 pKa = 6.02LIDD168 pKa = 3.75SCWQKK173 pKa = 11.3GSIKK177 pKa = 9.94IGRR180 pKa = 11.84AEE182 pKa = 4.16ASSIAYY188 pKa = 6.41TVGYY192 pKa = 9.7IVSKK196 pKa = 10.28EE197 pKa = 3.75KK198 pKa = 9.66KK199 pKa = 8.19TCFKK203 pKa = 10.72LQSQGLGFDD212 pKa = 3.45YY213 pKa = 10.64FKK215 pKa = 11.11NLNSSYY221 pKa = 11.04VLSTGRR227 pKa = 11.84GKK229 pKa = 10.27EE230 pKa = 3.85LYY232 pKa = 10.19VRR234 pKa = 11.84LPRR237 pKa = 11.84YY238 pKa = 9.69LKK240 pKa = 10.28EE241 pKa = 3.99KK242 pKa = 10.62YY243 pKa = 9.78GLKK246 pKa = 10.24SEE248 pKa = 4.21FDD250 pKa = 3.49SVKK253 pKa = 10.86AEE255 pKa = 4.0RR256 pKa = 11.84EE257 pKa = 4.07WKK259 pKa = 10.57NKK261 pKa = 9.21VFGSGLDD268 pKa = 3.48EE269 pKa = 4.07EE270 pKa = 5.4DD271 pKa = 3.54YY272 pKa = 11.45RR273 pKa = 11.84DD274 pKa = 3.72FKK276 pKa = 11.34QYY278 pKa = 11.04LSEE281 pKa = 4.37HH282 pKa = 6.37KK283 pKa = 10.67LIVHH287 pKa = 7.16
Molecular weight: 33.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1422 |
83 |
576 |
203.1 |
22.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.767 ± 1.328 | 1.266 ± 0.454 |
6.188 ± 0.848 | 6.61 ± 1.08 |
4.852 ± 0.408 | 7.032 ± 0.672 |
1.758 ± 0.42 | 5.626 ± 0.558 |
6.681 ± 1.357 | 8.931 ± 0.978 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.039 ± 0.501 | 5.345 ± 0.668 |
3.586 ± 0.981 | 2.813 ± 0.197 |
4.36 ± 0.439 | 9.986 ± 1.701 |
5.556 ± 0.722 | 4.993 ± 0.613 |
1.055 ± 0.228 | 5.556 ± 0.751 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
