
Afifella marina DSM 2698
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Afifellaceae; Afifella; Afifella marina
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
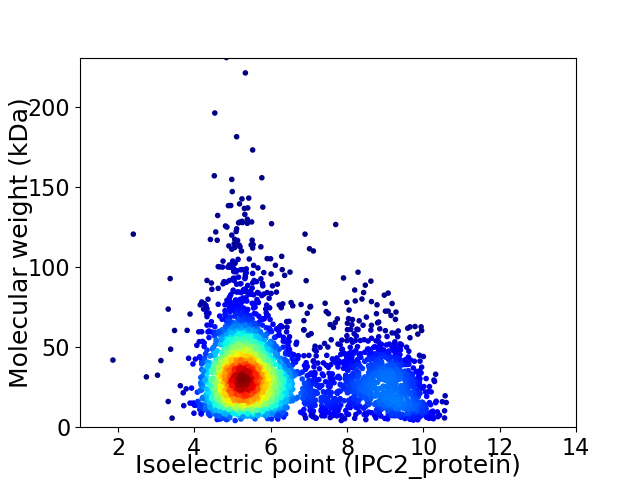
Virtual 2D-PAGE plot for 3662 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5NBI9|A0A1G5NBI9_AFIMA Pilus formation protein N terminal region OS=Afifella marina DSM 2698 OX=1120955 GN=SAMN03080610_01704 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84GTKK5 pKa = 9.69ILGRR9 pKa = 11.84AALAAAFLSIAGVAGAADD27 pKa = 4.47LPGQTYY33 pKa = 10.6EE34 pKa = 4.48PMAAPAPVAWSPWQIRR50 pKa = 11.84LRR52 pKa = 11.84ALAVVPHH59 pKa = 7.16DD60 pKa = 4.49SGSVDD65 pKa = 3.94QIPGSDD71 pKa = 3.17LSYY74 pKa = 10.91SNSVTPEE81 pKa = 3.35LDD83 pKa = 2.49ITYY86 pKa = 9.69FFTPNIAAEE95 pKa = 4.86LILGTTYY102 pKa = 11.41AEE104 pKa = 4.35VEE106 pKa = 3.96GDD108 pKa = 3.99GPISGLDD115 pKa = 4.2AGDD118 pKa = 3.48TWILPPTLTLQYY130 pKa = 10.68HH131 pKa = 5.57FTNFGAFKK139 pKa = 9.23PYY141 pKa = 10.25VGAGVNYY148 pKa = 8.09TFFYY152 pKa = 9.41GTEE155 pKa = 4.08NEE157 pKa = 4.47DD158 pKa = 3.49LAAFDD163 pKa = 4.26IDD165 pKa = 3.76NSFGLALQVGADD177 pKa = 3.61YY178 pKa = 10.19MFNEE182 pKa = 4.18HH183 pKa = 6.69WGLNLDD189 pKa = 3.85VKK191 pKa = 10.95KK192 pKa = 10.98LFLSADD198 pKa = 3.42WDD200 pKa = 3.89GSLADD205 pKa = 3.71GTYY208 pKa = 10.83VSGDD212 pKa = 3.31ADD214 pKa = 4.61LDD216 pKa = 3.49PWLIGAGITYY226 pKa = 10.3KK227 pKa = 10.78FF228 pKa = 3.65
MM1 pKa = 8.06RR2 pKa = 11.84GTKK5 pKa = 9.69ILGRR9 pKa = 11.84AALAAAFLSIAGVAGAADD27 pKa = 4.47LPGQTYY33 pKa = 10.6EE34 pKa = 4.48PMAAPAPVAWSPWQIRR50 pKa = 11.84LRR52 pKa = 11.84ALAVVPHH59 pKa = 7.16DD60 pKa = 4.49SGSVDD65 pKa = 3.94QIPGSDD71 pKa = 3.17LSYY74 pKa = 10.91SNSVTPEE81 pKa = 3.35LDD83 pKa = 2.49ITYY86 pKa = 9.69FFTPNIAAEE95 pKa = 4.86LILGTTYY102 pKa = 11.41AEE104 pKa = 4.35VEE106 pKa = 3.96GDD108 pKa = 3.99GPISGLDD115 pKa = 4.2AGDD118 pKa = 3.48TWILPPTLTLQYY130 pKa = 10.68HH131 pKa = 5.57FTNFGAFKK139 pKa = 9.23PYY141 pKa = 10.25VGAGVNYY148 pKa = 8.09TFFYY152 pKa = 9.41GTEE155 pKa = 4.08NEE157 pKa = 4.47DD158 pKa = 3.49LAAFDD163 pKa = 4.26IDD165 pKa = 3.76NSFGLALQVGADD177 pKa = 3.61YY178 pKa = 10.19MFNEE182 pKa = 4.18HH183 pKa = 6.69WGLNLDD189 pKa = 3.85VKK191 pKa = 10.95KK192 pKa = 10.98LFLSADD198 pKa = 3.42WDD200 pKa = 3.89GSLADD205 pKa = 3.71GTYY208 pKa = 10.83VSGDD212 pKa = 3.31ADD214 pKa = 4.61LDD216 pKa = 3.49PWLIGAGITYY226 pKa = 10.3KK227 pKa = 10.78FF228 pKa = 3.65
Molecular weight: 24.39 kDa
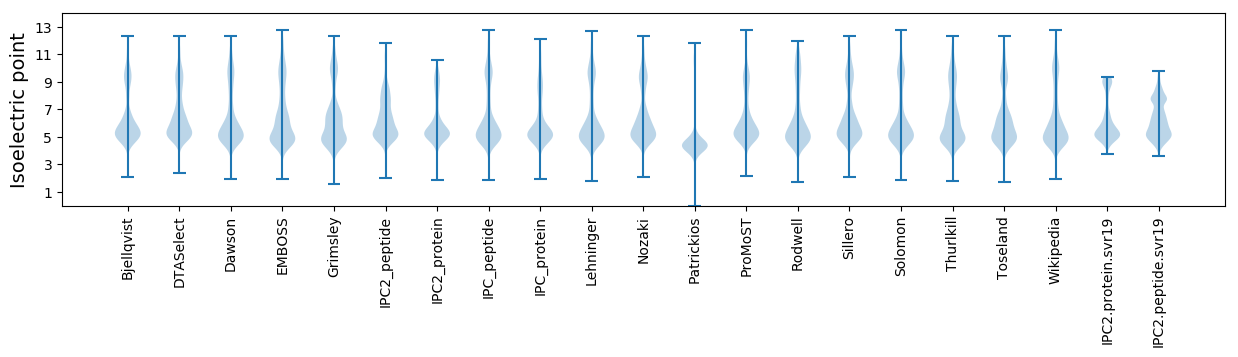
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5NRL6|A0A1G5NRL6_AFIMA Response regulator receiver domain-containing protein OS=Afifella marina DSM 2698 OX=1120955 GN=SAMN03080610_02542 PE=4 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84PRR4 pKa = 11.84KK5 pKa = 9.39RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SWFRR12 pKa = 11.84RR13 pKa = 11.84LVLTLLLVLCLPLFLIPIYY32 pKa = 10.64SIVNPPVTTLMLVKK46 pKa = 10.28RR47 pKa = 11.84LGGAPIEE54 pKa = 4.35RR55 pKa = 11.84RR56 pKa = 11.84WVPLDD61 pKa = 4.81DD62 pKa = 4.53ISPNLVNAVVMAEE75 pKa = 3.84DD76 pKa = 3.7AKK78 pKa = 10.5FCRR81 pKa = 11.84HH82 pKa = 6.04HH83 pKa = 7.11GIDD86 pKa = 3.99LGEE89 pKa = 4.74LRR91 pKa = 11.84AAWRR95 pKa = 11.84EE96 pKa = 3.97AQKK99 pKa = 10.8GGEE102 pKa = 4.03MRR104 pKa = 11.84GASTITMQTVKK115 pKa = 10.75NLFLWPSRR123 pKa = 11.84SYY125 pKa = 10.63VRR127 pKa = 11.84KK128 pKa = 9.69AVEE131 pKa = 4.46FPLALYY137 pKa = 10.92ADD139 pKa = 4.75FVLSKK144 pKa = 10.71RR145 pKa = 11.84RR146 pKa = 11.84ILEE149 pKa = 3.65IYY151 pKa = 10.15LNVVEE156 pKa = 4.72WDD158 pKa = 3.13RR159 pKa = 11.84GIYY162 pKa = 9.34GAEE165 pKa = 3.9AAAQHH170 pKa = 5.84YY171 pKa = 7.91FNRR174 pKa = 11.84PAASLTRR181 pKa = 11.84AEE183 pKa = 4.61GARR186 pKa = 11.84LAATLPAPDD195 pKa = 4.84LRR197 pKa = 11.84NPANPGPVTRR207 pKa = 11.84DD208 pKa = 3.14LARR211 pKa = 11.84LFARR215 pKa = 11.84RR216 pKa = 11.84AAAAGPHH223 pKa = 6.27IDD225 pKa = 4.22CLKK228 pKa = 10.5KK229 pKa = 10.7
MM1 pKa = 7.92RR2 pKa = 11.84PRR4 pKa = 11.84KK5 pKa = 9.39RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SWFRR12 pKa = 11.84RR13 pKa = 11.84LVLTLLLVLCLPLFLIPIYY32 pKa = 10.64SIVNPPVTTLMLVKK46 pKa = 10.28RR47 pKa = 11.84LGGAPIEE54 pKa = 4.35RR55 pKa = 11.84RR56 pKa = 11.84WVPLDD61 pKa = 4.81DD62 pKa = 4.53ISPNLVNAVVMAEE75 pKa = 3.84DD76 pKa = 3.7AKK78 pKa = 10.5FCRR81 pKa = 11.84HH82 pKa = 6.04HH83 pKa = 7.11GIDD86 pKa = 3.99LGEE89 pKa = 4.74LRR91 pKa = 11.84AAWRR95 pKa = 11.84EE96 pKa = 3.97AQKK99 pKa = 10.8GGEE102 pKa = 4.03MRR104 pKa = 11.84GASTITMQTVKK115 pKa = 10.75NLFLWPSRR123 pKa = 11.84SYY125 pKa = 10.63VRR127 pKa = 11.84KK128 pKa = 9.69AVEE131 pKa = 4.46FPLALYY137 pKa = 10.92ADD139 pKa = 4.75FVLSKK144 pKa = 10.71RR145 pKa = 11.84RR146 pKa = 11.84ILEE149 pKa = 3.65IYY151 pKa = 10.15LNVVEE156 pKa = 4.72WDD158 pKa = 3.13RR159 pKa = 11.84GIYY162 pKa = 9.34GAEE165 pKa = 3.9AAAQHH170 pKa = 5.84YY171 pKa = 7.91FNRR174 pKa = 11.84PAASLTRR181 pKa = 11.84AEE183 pKa = 4.61GARR186 pKa = 11.84LAATLPAPDD195 pKa = 4.84LRR197 pKa = 11.84NPANPGPVTRR207 pKa = 11.84DD208 pKa = 3.14LARR211 pKa = 11.84LFARR215 pKa = 11.84RR216 pKa = 11.84AAAAGPHH223 pKa = 6.27IDD225 pKa = 4.22CLKK228 pKa = 10.5KK229 pKa = 10.7
Molecular weight: 25.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1182367 |
39 |
2128 |
322.9 |
35.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.482 ± 0.057 | 0.799 ± 0.012 |
5.713 ± 0.037 | 6.619 ± 0.042 |
3.844 ± 0.029 | 8.522 ± 0.04 |
1.995 ± 0.017 | 5.148 ± 0.026 |
3.224 ± 0.03 | 10.242 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.019 | 2.42 ± 0.02 |
5.148 ± 0.033 | 2.87 ± 0.022 |
7.237 ± 0.039 | 5.517 ± 0.031 |
5.1 ± 0.025 | 7.355 ± 0.036 |
1.243 ± 0.016 | 2.151 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
