
Microviridae Fen2266_11
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
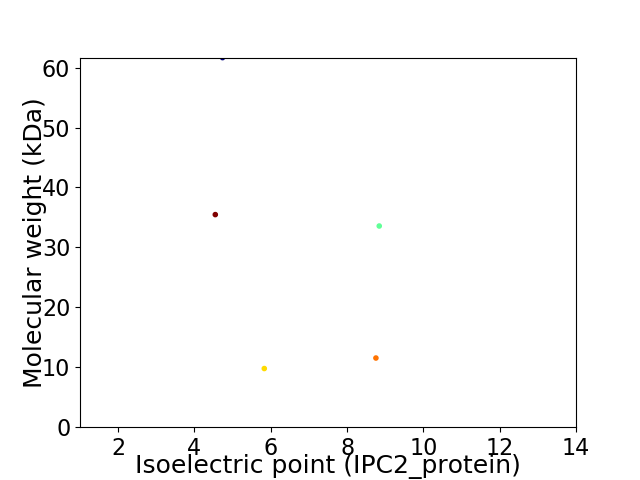
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UME2|A0A0G2UME2_9VIRU Uncharacterized protein OS=Microviridae Fen2266_11 OX=1655652 PE=4 SV=1
MM1 pKa = 7.9ADD3 pKa = 3.21SSDD6 pKa = 3.6PGLSIVAGAIPIVGGVISSLINNSAVDD33 pKa = 3.85ANNAANRR40 pKa = 11.84AFQVQQQQSQNEE52 pKa = 4.23YY53 pKa = 8.84NQAMWQEE60 pKa = 4.01QTDD63 pKa = 4.0YY64 pKa = 11.69NNPQNMMDD72 pKa = 3.61RR73 pKa = 11.84YY74 pKa = 9.45EE75 pKa = 4.25AAGLNPNLIYY85 pKa = 11.05GNATGNVAAQAPMEE99 pKa = 4.36AKK101 pKa = 9.88VDD103 pKa = 4.11YY104 pKa = 10.27VAQPHH109 pKa = 6.51PSVDD113 pKa = 3.38GSAMAAQAVQSANNTKK129 pKa = 10.33LVAAQSNNLQAQTTNTAEE147 pKa = 3.97DD148 pKa = 3.7TVLKK152 pKa = 10.64KK153 pKa = 10.41FVEE156 pKa = 4.47ALTSAEE162 pKa = 3.87ADD164 pKa = 3.44KK165 pKa = 11.07TSAEE169 pKa = 4.03ASFAGPTAQASIGNLLSSSFKK190 pKa = 10.77NQVDD194 pKa = 3.61AQTTSMLAPYY204 pKa = 9.19SANMQAAQTDD214 pKa = 4.0KK215 pKa = 11.43LKK217 pKa = 11.02ADD219 pKa = 3.76TNFTINDD226 pKa = 3.64NVRR229 pKa = 11.84QAALASTTIQEE240 pKa = 4.61AISRR244 pKa = 11.84IAMNAMTNAKK254 pKa = 9.5SAAEE258 pKa = 3.88IQQIRR263 pKa = 11.84ANTQLILGSKK273 pKa = 9.81ALQDD277 pKa = 3.49MDD279 pKa = 4.54LSMRR283 pKa = 11.84NAGMMPHH290 pKa = 7.28DD291 pKa = 4.52GFIQRR296 pKa = 11.84LLGGIGTWITGSNSAGGYY314 pKa = 9.18DD315 pKa = 4.13HH316 pKa = 7.64FDD318 pKa = 3.77GSPSTSSSVSPKK330 pKa = 10.64YY331 pKa = 10.65GDD333 pKa = 4.76PILGNN338 pKa = 3.7
MM1 pKa = 7.9ADD3 pKa = 3.21SSDD6 pKa = 3.6PGLSIVAGAIPIVGGVISSLINNSAVDD33 pKa = 3.85ANNAANRR40 pKa = 11.84AFQVQQQQSQNEE52 pKa = 4.23YY53 pKa = 8.84NQAMWQEE60 pKa = 4.01QTDD63 pKa = 4.0YY64 pKa = 11.69NNPQNMMDD72 pKa = 3.61RR73 pKa = 11.84YY74 pKa = 9.45EE75 pKa = 4.25AAGLNPNLIYY85 pKa = 11.05GNATGNVAAQAPMEE99 pKa = 4.36AKK101 pKa = 9.88VDD103 pKa = 4.11YY104 pKa = 10.27VAQPHH109 pKa = 6.51PSVDD113 pKa = 3.38GSAMAAQAVQSANNTKK129 pKa = 10.33LVAAQSNNLQAQTTNTAEE147 pKa = 3.97DD148 pKa = 3.7TVLKK152 pKa = 10.64KK153 pKa = 10.41FVEE156 pKa = 4.47ALTSAEE162 pKa = 3.87ADD164 pKa = 3.44KK165 pKa = 11.07TSAEE169 pKa = 4.03ASFAGPTAQASIGNLLSSSFKK190 pKa = 10.77NQVDD194 pKa = 3.61AQTTSMLAPYY204 pKa = 9.19SANMQAAQTDD214 pKa = 4.0KK215 pKa = 11.43LKK217 pKa = 11.02ADD219 pKa = 3.76TNFTINDD226 pKa = 3.64NVRR229 pKa = 11.84QAALASTTIQEE240 pKa = 4.61AISRR244 pKa = 11.84IAMNAMTNAKK254 pKa = 9.5SAAEE258 pKa = 3.88IQQIRR263 pKa = 11.84ANTQLILGSKK273 pKa = 9.81ALQDD277 pKa = 3.49MDD279 pKa = 4.54LSMRR283 pKa = 11.84NAGMMPHH290 pKa = 7.28DD291 pKa = 4.52GFIQRR296 pKa = 11.84LLGGIGTWITGSNSAGGYY314 pKa = 9.18DD315 pKa = 4.13HH316 pKa = 7.64FDD318 pKa = 3.77GSPSTSSSVSPKK330 pKa = 10.64YY331 pKa = 10.65GDD333 pKa = 4.76PILGNN338 pKa = 3.7
Molecular weight: 35.48 kDa
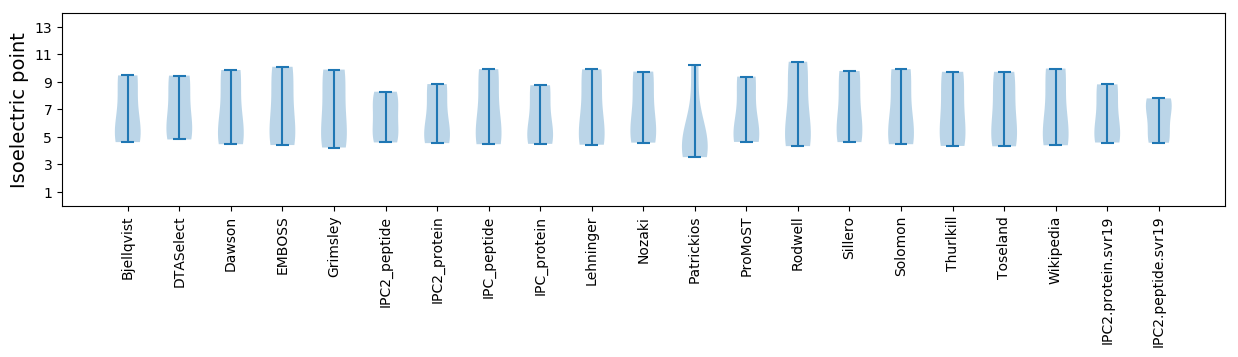
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UK25|A0A0G2UK25_9VIRU DNA pilot protein VP2 OS=Microviridae Fen2266_11 OX=1655652 PE=4 SV=1
MM1 pKa = 7.52ACMFPFFVDD10 pKa = 4.64DD11 pKa = 4.08VPVPCGKK18 pKa = 10.4CPACLVRR25 pKa = 11.84RR26 pKa = 11.84TNNWVFRR33 pKa = 11.84IMQEE37 pKa = 3.77EE38 pKa = 4.27RR39 pKa = 11.84HH40 pKa = 5.96SVSSLWITLTYY51 pKa = 11.0DD52 pKa = 3.34NYY54 pKa = 11.0HH55 pKa = 6.58LPISPNGFATLCKK68 pKa = 9.81RR69 pKa = 11.84DD70 pKa = 3.33VQLFFKK76 pKa = 10.39RR77 pKa = 11.84YY78 pKa = 8.8RR79 pKa = 11.84KK80 pKa = 9.96ALGKK84 pKa = 9.77DD85 pKa = 3.09HH86 pKa = 7.39PKK88 pKa = 9.92IKK90 pKa = 10.6YY91 pKa = 6.96YY92 pKa = 11.09ACGEE96 pKa = 4.24YY97 pKa = 10.43GSKK100 pKa = 10.06GQRR103 pKa = 11.84PHH105 pKa = 5.31YY106 pKa = 9.79HH107 pKa = 6.54LLIINGVDD115 pKa = 2.92RR116 pKa = 11.84CLASSWRR123 pKa = 11.84DD124 pKa = 3.35SEE126 pKa = 4.49GVQIGDD132 pKa = 3.68IYY134 pKa = 10.83IDD136 pKa = 4.04DD137 pKa = 4.36RR138 pKa = 11.84PLNSSAIGYY147 pKa = 6.58TVGYY151 pKa = 8.36MNKK154 pKa = 8.62TKK156 pKa = 9.38ITGRR160 pKa = 11.84YY161 pKa = 7.41QRR163 pKa = 11.84DD164 pKa = 3.42DD165 pKa = 3.6RR166 pKa = 11.84VSEE169 pKa = 3.99FALMSKK175 pKa = 10.61KK176 pKa = 9.79MGLSYY181 pKa = 9.36LTPEE185 pKa = 3.77VVAWHH190 pKa = 6.5KK191 pKa = 11.47ADD193 pKa = 3.75LSRR196 pKa = 11.84NYY198 pKa = 10.14VILPGGVKK206 pKa = 10.04SALPRR211 pKa = 11.84YY212 pKa = 8.58FADD215 pKa = 5.4RR216 pKa = 11.84IYY218 pKa = 11.23SDD220 pKa = 3.47EE221 pKa = 4.41DD222 pKa = 3.31KK223 pKa = 10.52EE224 pKa = 4.22ARR226 pKa = 11.84IAILKK231 pKa = 10.11LLFVKK236 pKa = 10.55LLSLNMMTMFVNLVMLMDD254 pKa = 4.93LLSLKK259 pKa = 8.94MRR261 pKa = 11.84PGMLQLLTFKK271 pKa = 10.34KK272 pKa = 10.45RR273 pKa = 11.84LMAVSTYY280 pKa = 11.2DD281 pKa = 3.75KK282 pKa = 10.92NFCAFGAYY290 pKa = 9.23HH291 pKa = 7.11
MM1 pKa = 7.52ACMFPFFVDD10 pKa = 4.64DD11 pKa = 4.08VPVPCGKK18 pKa = 10.4CPACLVRR25 pKa = 11.84RR26 pKa = 11.84TNNWVFRR33 pKa = 11.84IMQEE37 pKa = 3.77EE38 pKa = 4.27RR39 pKa = 11.84HH40 pKa = 5.96SVSSLWITLTYY51 pKa = 11.0DD52 pKa = 3.34NYY54 pKa = 11.0HH55 pKa = 6.58LPISPNGFATLCKK68 pKa = 9.81RR69 pKa = 11.84DD70 pKa = 3.33VQLFFKK76 pKa = 10.39RR77 pKa = 11.84YY78 pKa = 8.8RR79 pKa = 11.84KK80 pKa = 9.96ALGKK84 pKa = 9.77DD85 pKa = 3.09HH86 pKa = 7.39PKK88 pKa = 9.92IKK90 pKa = 10.6YY91 pKa = 6.96YY92 pKa = 11.09ACGEE96 pKa = 4.24YY97 pKa = 10.43GSKK100 pKa = 10.06GQRR103 pKa = 11.84PHH105 pKa = 5.31YY106 pKa = 9.79HH107 pKa = 6.54LLIINGVDD115 pKa = 2.92RR116 pKa = 11.84CLASSWRR123 pKa = 11.84DD124 pKa = 3.35SEE126 pKa = 4.49GVQIGDD132 pKa = 3.68IYY134 pKa = 10.83IDD136 pKa = 4.04DD137 pKa = 4.36RR138 pKa = 11.84PLNSSAIGYY147 pKa = 6.58TVGYY151 pKa = 8.36MNKK154 pKa = 8.62TKK156 pKa = 9.38ITGRR160 pKa = 11.84YY161 pKa = 7.41QRR163 pKa = 11.84DD164 pKa = 3.42DD165 pKa = 3.6RR166 pKa = 11.84VSEE169 pKa = 3.99FALMSKK175 pKa = 10.61KK176 pKa = 9.79MGLSYY181 pKa = 9.36LTPEE185 pKa = 3.77VVAWHH190 pKa = 6.5KK191 pKa = 11.47ADD193 pKa = 3.75LSRR196 pKa = 11.84NYY198 pKa = 10.14VILPGGVKK206 pKa = 10.04SALPRR211 pKa = 11.84YY212 pKa = 8.58FADD215 pKa = 5.4RR216 pKa = 11.84IYY218 pKa = 11.23SDD220 pKa = 3.47EE221 pKa = 4.41DD222 pKa = 3.31KK223 pKa = 10.52EE224 pKa = 4.22ARR226 pKa = 11.84IAILKK231 pKa = 10.11LLFVKK236 pKa = 10.55LLSLNMMTMFVNLVMLMDD254 pKa = 4.93LLSLKK259 pKa = 8.94MRR261 pKa = 11.84PGMLQLLTFKK271 pKa = 10.34KK272 pKa = 10.45RR273 pKa = 11.84LMAVSTYY280 pKa = 11.2DD281 pKa = 3.75KK282 pKa = 10.92NFCAFGAYY290 pKa = 9.23HH291 pKa = 7.11
Molecular weight: 33.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1373 |
88 |
554 |
274.6 |
30.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.124 ± 2.017 | 1.092 ± 0.541 |
6.555 ± 0.359 | 4.224 ± 0.649 |
4.006 ± 0.691 | 6.118 ± 0.747 |
2.112 ± 0.486 | 5.317 ± 0.089 |
4.807 ± 1.558 | 8.23 ± 1.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.496 ± 0.59 | 5.681 ± 1.201 |
5.171 ± 0.749 | 5.025 ± 1.357 |
4.443 ± 1.05 | 6.773 ± 1.033 |
5.899 ± 0.972 | 6.045 ± 0.515 |
1.311 ± 0.44 | 3.569 ± 0.757 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
