
Yichang Insect virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Goukovirus; Yichang insect goukovirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
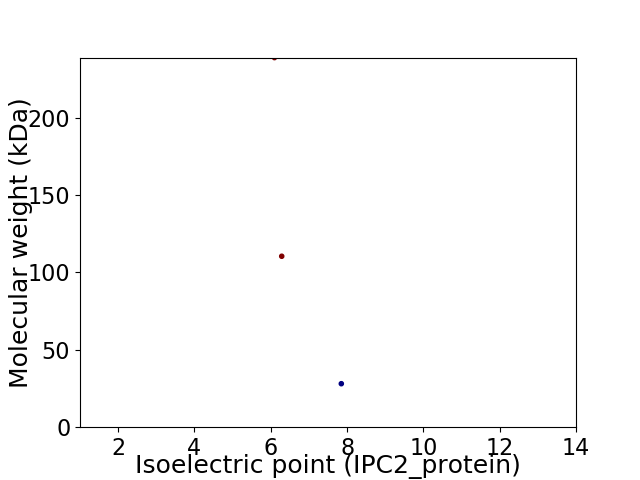
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
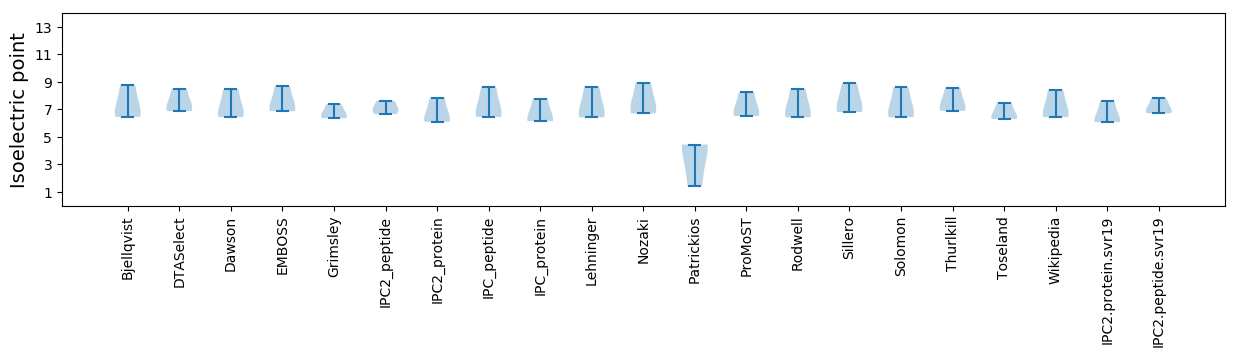
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXZ2|A0A0B5KXZ2_9VIRU Replicase OS=Yichang Insect virus OX=1608144 GN=L PE=4 SV=1
MM1 pKa = 7.09STRR4 pKa = 11.84TTGTYY9 pKa = 10.5PLVQDD14 pKa = 3.55MLKK17 pKa = 10.76ARR19 pKa = 11.84VPNTNLTLACLLAIGLSGCGGGVGMAQAEE48 pKa = 4.41IAASVPYY55 pKa = 9.76GAHH58 pKa = 6.44EE59 pKa = 4.76GMTQAACPDD68 pKa = 3.58EE69 pKa = 5.24KK70 pKa = 11.1LLSNKK75 pKa = 8.68WCGDD79 pKa = 2.92IPSTVNTCQPRR90 pKa = 11.84GGFIAQYY97 pKa = 9.85QWGDD101 pKa = 2.81KK102 pKa = 10.82YY103 pKa = 8.67YY104 pKa = 8.09TCSRR108 pKa = 11.84EE109 pKa = 4.45VICSPGEE116 pKa = 3.97YY117 pKa = 10.26LADD120 pKa = 4.45DD121 pKa = 4.2CTCALLIKK129 pKa = 10.24DD130 pKa = 4.99CYY132 pKa = 10.85HH133 pKa = 6.69EE134 pKa = 4.44AQNCRR139 pKa = 11.84VPKK142 pKa = 9.03LTKK145 pKa = 10.29LGSGTCTEE153 pKa = 4.86KK154 pKa = 10.63DD155 pKa = 3.01WKK157 pKa = 11.1ANEE160 pKa = 4.13PQFCSIGSPGEE171 pKa = 3.93GCNPIGVMRR180 pKa = 11.84EE181 pKa = 3.53LAYY184 pKa = 11.0VVDD187 pKa = 4.19STGKK191 pKa = 10.18SRR193 pKa = 11.84WITDD197 pKa = 3.6LNIEE201 pKa = 4.19EE202 pKa = 4.2ALIYY206 pKa = 10.48DD207 pKa = 3.75KK208 pKa = 11.34SEE210 pKa = 4.01SFIQDD215 pKa = 2.97KK216 pKa = 10.61HH217 pKa = 7.35VSLSGPVRR225 pKa = 11.84GNSAYY230 pKa = 10.26CDD232 pKa = 3.49HH233 pKa = 7.13FKK235 pKa = 11.07DD236 pKa = 5.31LPNCYY241 pKa = 10.44ASIASEE247 pKa = 3.75NSPYY251 pKa = 10.47RR252 pKa = 11.84IFRR255 pKa = 11.84SNGAIPHH262 pKa = 6.0IHH264 pKa = 7.51LDD266 pKa = 3.65TCSSPVYY273 pKa = 10.2CYY275 pKa = 10.59GYY277 pKa = 8.72KK278 pKa = 8.03TTLVYY283 pKa = 8.38THH285 pKa = 6.85KK286 pKa = 8.33TTEE289 pKa = 4.3LKK291 pKa = 10.63SSCSGCVVSCHH302 pKa = 4.85NTGVHH307 pKa = 5.82IVTNLEE313 pKa = 4.21GKK315 pKa = 8.68SHH317 pKa = 5.92IQVCGRR323 pKa = 11.84AEE325 pKa = 4.11CQMLDD330 pKa = 3.74TNSKK334 pKa = 10.04ILDD337 pKa = 3.78LPRR340 pKa = 11.84SFHH343 pKa = 7.28SRR345 pKa = 11.84TTDD348 pKa = 2.68EE349 pKa = 4.89RR350 pKa = 11.84IRR352 pKa = 11.84VTIHH356 pKa = 6.18SEE358 pKa = 3.92DD359 pKa = 3.74RR360 pKa = 11.84LVSYY364 pKa = 11.12DD365 pKa = 4.03NFTACPVVPVCEE377 pKa = 5.55AIDD380 pKa = 3.58CTFCSDD386 pKa = 3.33HH387 pKa = 6.74LLNPTCYY394 pKa = 10.5SKK396 pKa = 11.35LDD398 pKa = 3.47WVLTLVFLYY407 pKa = 11.08VLMVLTGLALTCAIPIFRR425 pKa = 11.84VLWNLVSFMIKK436 pKa = 10.24LFWKK440 pKa = 5.9MTKK443 pKa = 9.56MMLRR447 pKa = 11.84MSGRR451 pKa = 11.84AAGRR455 pKa = 11.84AKK457 pKa = 10.35QFADD461 pKa = 3.89DD462 pKa = 4.93DD463 pKa = 4.95SIEE466 pKa = 4.31LRR468 pKa = 11.84PASIVRR474 pKa = 11.84EE475 pKa = 3.89PLINSRR481 pKa = 11.84PARR484 pKa = 11.84QNKK487 pKa = 9.84AGIRR491 pKa = 11.84LPAGLVIIMILLPLVVCKK509 pKa = 10.47APCSSVVVDD518 pKa = 3.67TVMTEE523 pKa = 3.64SCTLKK528 pKa = 10.54GDD530 pKa = 4.08SYY532 pKa = 11.12HH533 pKa = 7.13CSISSINEE541 pKa = 3.78VPLISYY547 pKa = 8.81DD548 pKa = 3.4QTTCLMYY555 pKa = 9.17LTPGGQTLGQLQITPLDD572 pKa = 3.41ISFKK576 pKa = 10.26CSKK579 pKa = 9.52QALYY583 pKa = 8.64HH584 pKa = 5.56TRR586 pKa = 11.84DD587 pKa = 3.26FEE589 pKa = 4.23MLSDD593 pKa = 3.61HH594 pKa = 6.62VVRR597 pKa = 11.84CPRR600 pKa = 11.84SGPCTEE606 pKa = 4.72DD607 pKa = 2.62WCHH610 pKa = 6.8SVTTDD615 pKa = 3.03TDD617 pKa = 3.84VPDD620 pKa = 4.35LPKK623 pKa = 10.18ISYY626 pKa = 9.01PSHH629 pKa = 5.88QSCKK633 pKa = 10.35LGEE636 pKa = 4.29ACWGNGCFFCTSSCHH651 pKa = 5.84VIRR654 pKa = 11.84YY655 pKa = 5.98YY656 pKa = 11.15ARR658 pKa = 11.84PKK660 pKa = 10.0TDD662 pKa = 3.59TIYY665 pKa = 11.05EE666 pKa = 4.35LYY668 pKa = 10.44SCPKK672 pKa = 8.47WDD674 pKa = 3.86PSGSFHH680 pKa = 6.97FLWEE684 pKa = 4.36STSGNTEE691 pKa = 3.82TTISLIHH698 pKa = 6.85GEE700 pKa = 4.35TKK702 pKa = 10.55SISSDD707 pKa = 3.01MSVTLDD713 pKa = 3.53VNIQSKK719 pKa = 10.79LPILSKK725 pKa = 9.89TFISDD730 pKa = 3.27GRR732 pKa = 11.84RR733 pKa = 11.84VALSDD738 pKa = 3.68SSFAGQPVAGQLGQVQCHH756 pKa = 5.08SAEE759 pKa = 4.3KK760 pKa = 10.27ASKK763 pKa = 10.14LQSCTMAPNICVCRR777 pKa = 11.84PEE779 pKa = 4.45QSDD782 pKa = 4.17DD783 pKa = 3.81GCDD786 pKa = 3.42CSQVFIDD793 pKa = 5.66RR794 pKa = 11.84IFSSSAVLPVNVEE807 pKa = 3.74NHH809 pKa = 6.4HH810 pKa = 7.89IEE812 pKa = 4.23MNRR815 pKa = 11.84DD816 pKa = 3.03TPYY819 pKa = 11.25LKK821 pKa = 9.88TPAFGSGHH829 pKa = 5.37IKK831 pKa = 10.07VRR833 pKa = 11.84SSMKK837 pKa = 10.56LIGTAVPAEE846 pKa = 4.26SCSVNLIKK854 pKa = 10.56ISGCYY859 pKa = 9.65SCVSGARR866 pKa = 11.84LSYY869 pKa = 10.12SCSTEE874 pKa = 3.5KK875 pKa = 10.56HH876 pKa = 5.04HH877 pKa = 6.79TVVASCYY884 pKa = 10.71GDD886 pKa = 2.61IHH888 pKa = 7.93LILEE892 pKa = 4.59CDD894 pKa = 3.15NSGVEE899 pKa = 4.0RR900 pKa = 11.84EE901 pKa = 4.31VRR903 pKa = 11.84FQSPTPLITLQCTTACSKK921 pKa = 10.37MGFSLSGTLVRR932 pKa = 11.84MAQPDD937 pKa = 3.88LSGQGHH943 pKa = 6.77INLVPGAAKK952 pKa = 10.18EE953 pKa = 4.18GAIEE957 pKa = 4.15WLRR960 pKa = 11.84QCWSLLGWFNMFWLVGMVSMVVLVLYY986 pKa = 8.75FCQRR990 pKa = 11.84RR991 pKa = 11.84ASPTHH996 pKa = 4.71IVKK999 pKa = 10.51YY1000 pKa = 10.08IKK1002 pKa = 10.63SSS1004 pKa = 3.08
MM1 pKa = 7.09STRR4 pKa = 11.84TTGTYY9 pKa = 10.5PLVQDD14 pKa = 3.55MLKK17 pKa = 10.76ARR19 pKa = 11.84VPNTNLTLACLLAIGLSGCGGGVGMAQAEE48 pKa = 4.41IAASVPYY55 pKa = 9.76GAHH58 pKa = 6.44EE59 pKa = 4.76GMTQAACPDD68 pKa = 3.58EE69 pKa = 5.24KK70 pKa = 11.1LLSNKK75 pKa = 8.68WCGDD79 pKa = 2.92IPSTVNTCQPRR90 pKa = 11.84GGFIAQYY97 pKa = 9.85QWGDD101 pKa = 2.81KK102 pKa = 10.82YY103 pKa = 8.67YY104 pKa = 8.09TCSRR108 pKa = 11.84EE109 pKa = 4.45VICSPGEE116 pKa = 3.97YY117 pKa = 10.26LADD120 pKa = 4.45DD121 pKa = 4.2CTCALLIKK129 pKa = 10.24DD130 pKa = 4.99CYY132 pKa = 10.85HH133 pKa = 6.69EE134 pKa = 4.44AQNCRR139 pKa = 11.84VPKK142 pKa = 9.03LTKK145 pKa = 10.29LGSGTCTEE153 pKa = 4.86KK154 pKa = 10.63DD155 pKa = 3.01WKK157 pKa = 11.1ANEE160 pKa = 4.13PQFCSIGSPGEE171 pKa = 3.93GCNPIGVMRR180 pKa = 11.84EE181 pKa = 3.53LAYY184 pKa = 11.0VVDD187 pKa = 4.19STGKK191 pKa = 10.18SRR193 pKa = 11.84WITDD197 pKa = 3.6LNIEE201 pKa = 4.19EE202 pKa = 4.2ALIYY206 pKa = 10.48DD207 pKa = 3.75KK208 pKa = 11.34SEE210 pKa = 4.01SFIQDD215 pKa = 2.97KK216 pKa = 10.61HH217 pKa = 7.35VSLSGPVRR225 pKa = 11.84GNSAYY230 pKa = 10.26CDD232 pKa = 3.49HH233 pKa = 7.13FKK235 pKa = 11.07DD236 pKa = 5.31LPNCYY241 pKa = 10.44ASIASEE247 pKa = 3.75NSPYY251 pKa = 10.47RR252 pKa = 11.84IFRR255 pKa = 11.84SNGAIPHH262 pKa = 6.0IHH264 pKa = 7.51LDD266 pKa = 3.65TCSSPVYY273 pKa = 10.2CYY275 pKa = 10.59GYY277 pKa = 8.72KK278 pKa = 8.03TTLVYY283 pKa = 8.38THH285 pKa = 6.85KK286 pKa = 8.33TTEE289 pKa = 4.3LKK291 pKa = 10.63SSCSGCVVSCHH302 pKa = 4.85NTGVHH307 pKa = 5.82IVTNLEE313 pKa = 4.21GKK315 pKa = 8.68SHH317 pKa = 5.92IQVCGRR323 pKa = 11.84AEE325 pKa = 4.11CQMLDD330 pKa = 3.74TNSKK334 pKa = 10.04ILDD337 pKa = 3.78LPRR340 pKa = 11.84SFHH343 pKa = 7.28SRR345 pKa = 11.84TTDD348 pKa = 2.68EE349 pKa = 4.89RR350 pKa = 11.84IRR352 pKa = 11.84VTIHH356 pKa = 6.18SEE358 pKa = 3.92DD359 pKa = 3.74RR360 pKa = 11.84LVSYY364 pKa = 11.12DD365 pKa = 4.03NFTACPVVPVCEE377 pKa = 5.55AIDD380 pKa = 3.58CTFCSDD386 pKa = 3.33HH387 pKa = 6.74LLNPTCYY394 pKa = 10.5SKK396 pKa = 11.35LDD398 pKa = 3.47WVLTLVFLYY407 pKa = 11.08VLMVLTGLALTCAIPIFRR425 pKa = 11.84VLWNLVSFMIKK436 pKa = 10.24LFWKK440 pKa = 5.9MTKK443 pKa = 9.56MMLRR447 pKa = 11.84MSGRR451 pKa = 11.84AAGRR455 pKa = 11.84AKK457 pKa = 10.35QFADD461 pKa = 3.89DD462 pKa = 4.93DD463 pKa = 4.95SIEE466 pKa = 4.31LRR468 pKa = 11.84PASIVRR474 pKa = 11.84EE475 pKa = 3.89PLINSRR481 pKa = 11.84PARR484 pKa = 11.84QNKK487 pKa = 9.84AGIRR491 pKa = 11.84LPAGLVIIMILLPLVVCKK509 pKa = 10.47APCSSVVVDD518 pKa = 3.67TVMTEE523 pKa = 3.64SCTLKK528 pKa = 10.54GDD530 pKa = 4.08SYY532 pKa = 11.12HH533 pKa = 7.13CSISSINEE541 pKa = 3.78VPLISYY547 pKa = 8.81DD548 pKa = 3.4QTTCLMYY555 pKa = 9.17LTPGGQTLGQLQITPLDD572 pKa = 3.41ISFKK576 pKa = 10.26CSKK579 pKa = 9.52QALYY583 pKa = 8.64HH584 pKa = 5.56TRR586 pKa = 11.84DD587 pKa = 3.26FEE589 pKa = 4.23MLSDD593 pKa = 3.61HH594 pKa = 6.62VVRR597 pKa = 11.84CPRR600 pKa = 11.84SGPCTEE606 pKa = 4.72DD607 pKa = 2.62WCHH610 pKa = 6.8SVTTDD615 pKa = 3.03TDD617 pKa = 3.84VPDD620 pKa = 4.35LPKK623 pKa = 10.18ISYY626 pKa = 9.01PSHH629 pKa = 5.88QSCKK633 pKa = 10.35LGEE636 pKa = 4.29ACWGNGCFFCTSSCHH651 pKa = 5.84VIRR654 pKa = 11.84YY655 pKa = 5.98YY656 pKa = 11.15ARR658 pKa = 11.84PKK660 pKa = 10.0TDD662 pKa = 3.59TIYY665 pKa = 11.05EE666 pKa = 4.35LYY668 pKa = 10.44SCPKK672 pKa = 8.47WDD674 pKa = 3.86PSGSFHH680 pKa = 6.97FLWEE684 pKa = 4.36STSGNTEE691 pKa = 3.82TTISLIHH698 pKa = 6.85GEE700 pKa = 4.35TKK702 pKa = 10.55SISSDD707 pKa = 3.01MSVTLDD713 pKa = 3.53VNIQSKK719 pKa = 10.79LPILSKK725 pKa = 9.89TFISDD730 pKa = 3.27GRR732 pKa = 11.84RR733 pKa = 11.84VALSDD738 pKa = 3.68SSFAGQPVAGQLGQVQCHH756 pKa = 5.08SAEE759 pKa = 4.3KK760 pKa = 10.27ASKK763 pKa = 10.14LQSCTMAPNICVCRR777 pKa = 11.84PEE779 pKa = 4.45QSDD782 pKa = 4.17DD783 pKa = 3.81GCDD786 pKa = 3.42CSQVFIDD793 pKa = 5.66RR794 pKa = 11.84IFSSSAVLPVNVEE807 pKa = 3.74NHH809 pKa = 6.4HH810 pKa = 7.89IEE812 pKa = 4.23MNRR815 pKa = 11.84DD816 pKa = 3.03TPYY819 pKa = 11.25LKK821 pKa = 9.88TPAFGSGHH829 pKa = 5.37IKK831 pKa = 10.07VRR833 pKa = 11.84SSMKK837 pKa = 10.56LIGTAVPAEE846 pKa = 4.26SCSVNLIKK854 pKa = 10.56ISGCYY859 pKa = 9.65SCVSGARR866 pKa = 11.84LSYY869 pKa = 10.12SCSTEE874 pKa = 3.5KK875 pKa = 10.56HH876 pKa = 5.04HH877 pKa = 6.79TVVASCYY884 pKa = 10.71GDD886 pKa = 2.61IHH888 pKa = 7.93LILEE892 pKa = 4.59CDD894 pKa = 3.15NSGVEE899 pKa = 4.0RR900 pKa = 11.84EE901 pKa = 4.31VRR903 pKa = 11.84FQSPTPLITLQCTTACSKK921 pKa = 10.37MGFSLSGTLVRR932 pKa = 11.84MAQPDD937 pKa = 3.88LSGQGHH943 pKa = 6.77INLVPGAAKK952 pKa = 10.18EE953 pKa = 4.18GAIEE957 pKa = 4.15WLRR960 pKa = 11.84QCWSLLGWFNMFWLVGMVSMVVLVLYY986 pKa = 8.75FCQRR990 pKa = 11.84RR991 pKa = 11.84ASPTHH996 pKa = 4.71IVKK999 pKa = 10.51YY1000 pKa = 10.08IKK1002 pKa = 10.63SSS1004 pKa = 3.08
Molecular weight: 110.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KKQ5|A0A0B5KKQ5_9VIRU Glycoprotein OS=Yichang Insect virus OX=1608144 GN=G PE=4 SV=1
MM1 pKa = 7.34TGVADD6 pKa = 3.83EE7 pKa = 4.64ARR9 pKa = 11.84AILEE13 pKa = 4.18SLSTEE18 pKa = 4.25TVFGGMNLDD27 pKa = 3.66VAEE30 pKa = 4.37MEE32 pKa = 4.48YY33 pKa = 10.65QGFSPDD39 pKa = 3.42LLIRR43 pKa = 11.84AMWEE47 pKa = 3.65LATKK51 pKa = 10.33KK52 pKa = 10.74GILADD57 pKa = 3.82SFKK60 pKa = 11.11KK61 pKa = 10.32DD62 pKa = 3.64LRR64 pKa = 11.84TMAVMGLMRR73 pKa = 11.84GSNIRR78 pKa = 11.84AIKK81 pKa = 10.34AKK83 pKa = 9.47STMEE87 pKa = 3.99AKK89 pKa = 10.44AQLTSWEE96 pKa = 4.21TTYY99 pKa = 11.45GLKK102 pKa = 10.24SGKK105 pKa = 7.53PTSSTTVTLVRR116 pKa = 11.84VSACLARR123 pKa = 11.84PMSLAMYY130 pKa = 9.18NGSLAISGAVPAEE143 pKa = 3.74AVVPGFPKK151 pKa = 10.69GMALSSFGSLIPGVDD166 pKa = 4.15TIPDD170 pKa = 3.7EE171 pKa = 4.56VCRR174 pKa = 11.84TLADD178 pKa = 2.91AFMYY182 pKa = 9.8HH183 pKa = 5.79QYY185 pKa = 11.38KK186 pKa = 9.29FDD188 pKa = 3.81KK189 pKa = 10.82VINSNTNRR197 pKa = 11.84EE198 pKa = 3.74TSIANIKK205 pKa = 10.15QYY207 pKa = 8.91TAIQMRR213 pKa = 11.84SNLYY217 pKa = 10.41DD218 pKa = 3.31SNARR222 pKa = 11.84VNHH225 pKa = 6.4CCTINILVKK234 pKa = 10.8DD235 pKa = 4.09GSTYY239 pKa = 10.92KK240 pKa = 10.75LSPSCSTALTTAAEE254 pKa = 4.42LFRR257 pKa = 11.84ALSS260 pKa = 3.55
MM1 pKa = 7.34TGVADD6 pKa = 3.83EE7 pKa = 4.64ARR9 pKa = 11.84AILEE13 pKa = 4.18SLSTEE18 pKa = 4.25TVFGGMNLDD27 pKa = 3.66VAEE30 pKa = 4.37MEE32 pKa = 4.48YY33 pKa = 10.65QGFSPDD39 pKa = 3.42LLIRR43 pKa = 11.84AMWEE47 pKa = 3.65LATKK51 pKa = 10.33KK52 pKa = 10.74GILADD57 pKa = 3.82SFKK60 pKa = 11.11KK61 pKa = 10.32DD62 pKa = 3.64LRR64 pKa = 11.84TMAVMGLMRR73 pKa = 11.84GSNIRR78 pKa = 11.84AIKK81 pKa = 10.34AKK83 pKa = 9.47STMEE87 pKa = 3.99AKK89 pKa = 10.44AQLTSWEE96 pKa = 4.21TTYY99 pKa = 11.45GLKK102 pKa = 10.24SGKK105 pKa = 7.53PTSSTTVTLVRR116 pKa = 11.84VSACLARR123 pKa = 11.84PMSLAMYY130 pKa = 9.18NGSLAISGAVPAEE143 pKa = 3.74AVVPGFPKK151 pKa = 10.69GMALSSFGSLIPGVDD166 pKa = 4.15TIPDD170 pKa = 3.7EE171 pKa = 4.56VCRR174 pKa = 11.84TLADD178 pKa = 2.91AFMYY182 pKa = 9.8HH183 pKa = 5.79QYY185 pKa = 11.38KK186 pKa = 9.29FDD188 pKa = 3.81KK189 pKa = 10.82VINSNTNRR197 pKa = 11.84EE198 pKa = 3.74TSIANIKK205 pKa = 10.15QYY207 pKa = 8.91TAIQMRR213 pKa = 11.84SNLYY217 pKa = 10.41DD218 pKa = 3.31SNARR222 pKa = 11.84VNHH225 pKa = 6.4CCTINILVKK234 pKa = 10.8DD235 pKa = 4.09GSTYY239 pKa = 10.92KK240 pKa = 10.75LSPSCSTALTTAAEE254 pKa = 4.42LFRR257 pKa = 11.84ALSS260 pKa = 3.55
Molecular weight: 28.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3353 |
260 |
2089 |
1117.7 |
125.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.875 ± 1.039 | 2.654 ± 1.435 |
5.637 ± 0.451 | 6.024 ± 0.922 |
3.639 ± 0.374 | 5.637 ± 0.407 |
2.058 ± 0.429 | 5.607 ± 0.2 |
5.577 ± 0.431 | 9.514 ± 0.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.668 ± 0.523 | 3.698 ± 0.225 |
4.384 ± 0.318 | 3.161 ± 0.242 |
5.786 ± 0.755 | 9.514 ± 0.45 |
6.203 ± 0.696 | 6.531 ± 0.216 |
1.64 ± 0.201 | 3.191 ± 0.083 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
