
Flavobacteriaceae bacterium YIM 102668
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
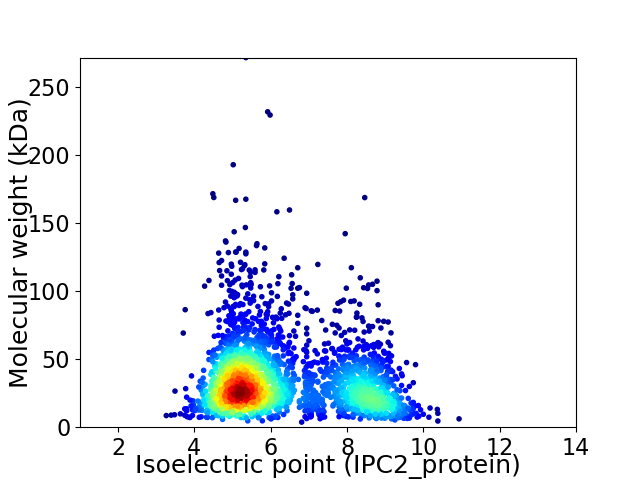
Virtual 2D-PAGE plot for 2667 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L9M067|A0A3L9M067_9FLAO DUF262 domain-containing protein OS=Flavobacteriaceae bacterium YIM 102668 OX=2479857 GN=EAH69_13805 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 10.38KK3 pKa = 8.47ITLSLFAAMATSFAAAQVDD22 pKa = 4.05QLSNPDD28 pKa = 3.27VSGGNNNYY36 pKa = 10.27SSAEE40 pKa = 3.82QYY42 pKa = 11.33GDD44 pKa = 3.51RR45 pKa = 11.84NQISVLQEE53 pKa = 4.23GNDD56 pKa = 3.55NTSDD60 pKa = 3.2ILQDD64 pKa = 3.45GNRR67 pKa = 11.84NMNTTVSTGNSNDD80 pKa = 3.46SKK82 pKa = 11.47VEE84 pKa = 3.69QFGNSNVGNQTQTGDD99 pKa = 3.69LNSARR104 pKa = 11.84LLQEE108 pKa = 3.89GLSNRR113 pKa = 11.84SEE115 pKa = 4.0LVQTGNGNGSEE126 pKa = 4.09SFQYY130 pKa = 9.52GTLNRR135 pKa = 11.84VTTTQTGNEE144 pKa = 3.97NYY146 pKa = 7.96ATNYY150 pKa = 8.5QDD152 pKa = 4.81GVINSIAQTQNGNNNFIGAGQFGVGNSATQSQDD185 pKa = 2.73GNSNYY190 pKa = 10.77AGTQQDD196 pKa = 3.46GNANLVLQTQVGDD209 pKa = 3.79YY210 pKa = 10.87NEE212 pKa = 4.64ADD214 pKa = 3.44AAQEE218 pKa = 4.18GSGNLSVQTQALVANTANSFQDD240 pKa = 3.46GNFNEE245 pKa = 4.97SYY247 pKa = 8.9QTQTGFGQSALVEE260 pKa = 3.95QDD262 pKa = 2.92GNRR265 pKa = 11.84NIATQDD271 pKa = 3.01QLGYY275 pKa = 10.32FNEE278 pKa = 4.37AVASQTGNRR287 pKa = 11.84NEE289 pKa = 4.03SSQTQEE295 pKa = 3.03GWANEE300 pKa = 4.18AYY302 pKa = 10.71VMQDD306 pKa = 2.52GDD308 pKa = 3.91RR309 pKa = 11.84NNAVQSQNGLLNYY322 pKa = 9.94AISEE326 pKa = 4.11QTGNRR331 pKa = 11.84NEE333 pKa = 4.13SNQFQEE339 pKa = 4.44GLGNGAAIVQDD350 pKa = 3.86GNRR353 pKa = 11.84NSASQLQIGDD363 pKa = 3.71GNGAEE368 pKa = 4.46VYY370 pKa = 11.02QMGNEE375 pKa = 3.88NLASQIQLGDD385 pKa = 3.28GNYY388 pKa = 11.16ALIEE392 pKa = 3.94QAGDD396 pKa = 3.62GNSVFQGQGGDD407 pKa = 3.38NNIAYY412 pKa = 9.98SSQGGDD418 pKa = 3.53EE419 pKa = 4.36NSSSQLQIGDD429 pKa = 3.67GNIALGLQIGDD440 pKa = 3.42NNEE443 pKa = 3.98FVQNQYY449 pKa = 10.82GFEE452 pKa = 4.06NVAQTVQFGVDD463 pKa = 3.11NYY465 pKa = 11.78AHH467 pKa = 5.65VTQDD471 pKa = 3.05GSSNMADD478 pKa = 3.17QYY480 pKa = 11.58QEE482 pKa = 3.93GVNNVAAIEE491 pKa = 4.0QIGNGNSATQTQDD504 pKa = 3.05DD505 pKa = 4.75FGRR508 pKa = 11.84APNWNNVGGNDD519 pKa = 2.82NYY521 pKa = 11.49AYY523 pKa = 10.47VRR525 pKa = 11.84QEE527 pKa = 3.31GDD529 pKa = 3.58GNVSIQGQNGSNNEE543 pKa = 3.75IASLQRR549 pKa = 11.84GDD551 pKa = 3.47ANYY554 pKa = 10.47VEE556 pKa = 5.15TYY558 pKa = 10.63QDD560 pKa = 3.16GFDD563 pKa = 3.52NFGVSVQLGNNNEE576 pKa = 3.7IRR578 pKa = 11.84MGQHH582 pKa = 6.44GDD584 pKa = 3.22SNMADD589 pKa = 2.95TWQEE593 pKa = 3.95GNGNIANSTQIGLEE607 pKa = 3.68NYY609 pKa = 7.66TFNTQIGNEE618 pKa = 4.16NTLTTYY624 pKa = 11.0QNGAANQSYY633 pKa = 10.21IIQDD637 pKa = 3.48GNRR640 pKa = 11.84NNAEE644 pKa = 3.95VIQRR648 pKa = 11.84GRR650 pKa = 3.22
MM1 pKa = 7.55KK2 pKa = 10.38KK3 pKa = 8.47ITLSLFAAMATSFAAAQVDD22 pKa = 4.05QLSNPDD28 pKa = 3.27VSGGNNNYY36 pKa = 10.27SSAEE40 pKa = 3.82QYY42 pKa = 11.33GDD44 pKa = 3.51RR45 pKa = 11.84NQISVLQEE53 pKa = 4.23GNDD56 pKa = 3.55NTSDD60 pKa = 3.2ILQDD64 pKa = 3.45GNRR67 pKa = 11.84NMNTTVSTGNSNDD80 pKa = 3.46SKK82 pKa = 11.47VEE84 pKa = 3.69QFGNSNVGNQTQTGDD99 pKa = 3.69LNSARR104 pKa = 11.84LLQEE108 pKa = 3.89GLSNRR113 pKa = 11.84SEE115 pKa = 4.0LVQTGNGNGSEE126 pKa = 4.09SFQYY130 pKa = 9.52GTLNRR135 pKa = 11.84VTTTQTGNEE144 pKa = 3.97NYY146 pKa = 7.96ATNYY150 pKa = 8.5QDD152 pKa = 4.81GVINSIAQTQNGNNNFIGAGQFGVGNSATQSQDD185 pKa = 2.73GNSNYY190 pKa = 10.77AGTQQDD196 pKa = 3.46GNANLVLQTQVGDD209 pKa = 3.79YY210 pKa = 10.87NEE212 pKa = 4.64ADD214 pKa = 3.44AAQEE218 pKa = 4.18GSGNLSVQTQALVANTANSFQDD240 pKa = 3.46GNFNEE245 pKa = 4.97SYY247 pKa = 8.9QTQTGFGQSALVEE260 pKa = 3.95QDD262 pKa = 2.92GNRR265 pKa = 11.84NIATQDD271 pKa = 3.01QLGYY275 pKa = 10.32FNEE278 pKa = 4.37AVASQTGNRR287 pKa = 11.84NEE289 pKa = 4.03SSQTQEE295 pKa = 3.03GWANEE300 pKa = 4.18AYY302 pKa = 10.71VMQDD306 pKa = 2.52GDD308 pKa = 3.91RR309 pKa = 11.84NNAVQSQNGLLNYY322 pKa = 9.94AISEE326 pKa = 4.11QTGNRR331 pKa = 11.84NEE333 pKa = 4.13SNQFQEE339 pKa = 4.44GLGNGAAIVQDD350 pKa = 3.86GNRR353 pKa = 11.84NSASQLQIGDD363 pKa = 3.71GNGAEE368 pKa = 4.46VYY370 pKa = 11.02QMGNEE375 pKa = 3.88NLASQIQLGDD385 pKa = 3.28GNYY388 pKa = 11.16ALIEE392 pKa = 3.94QAGDD396 pKa = 3.62GNSVFQGQGGDD407 pKa = 3.38NNIAYY412 pKa = 9.98SSQGGDD418 pKa = 3.53EE419 pKa = 4.36NSSSQLQIGDD429 pKa = 3.67GNIALGLQIGDD440 pKa = 3.42NNEE443 pKa = 3.98FVQNQYY449 pKa = 10.82GFEE452 pKa = 4.06NVAQTVQFGVDD463 pKa = 3.11NYY465 pKa = 11.78AHH467 pKa = 5.65VTQDD471 pKa = 3.05GSSNMADD478 pKa = 3.17QYY480 pKa = 11.58QEE482 pKa = 3.93GVNNVAAIEE491 pKa = 4.0QIGNGNSATQTQDD504 pKa = 3.05DD505 pKa = 4.75FGRR508 pKa = 11.84APNWNNVGGNDD519 pKa = 2.82NYY521 pKa = 11.49AYY523 pKa = 10.47VRR525 pKa = 11.84QEE527 pKa = 3.31GDD529 pKa = 3.58GNVSIQGQNGSNNEE543 pKa = 3.75IASLQRR549 pKa = 11.84GDD551 pKa = 3.47ANYY554 pKa = 10.47VEE556 pKa = 5.15TYY558 pKa = 10.63QDD560 pKa = 3.16GFDD563 pKa = 3.52NFGVSVQLGNNNEE576 pKa = 3.7IRR578 pKa = 11.84MGQHH582 pKa = 6.44GDD584 pKa = 3.22SNMADD589 pKa = 2.95TWQEE593 pKa = 3.95GNGNIANSTQIGLEE607 pKa = 3.68NYY609 pKa = 7.66TFNTQIGNEE618 pKa = 4.16NTLTTYY624 pKa = 11.0QNGAANQSYY633 pKa = 10.21IIQDD637 pKa = 3.48GNRR640 pKa = 11.84NNAEE644 pKa = 3.95VIQRR648 pKa = 11.84GRR650 pKa = 3.22
Molecular weight: 69.28 kDa
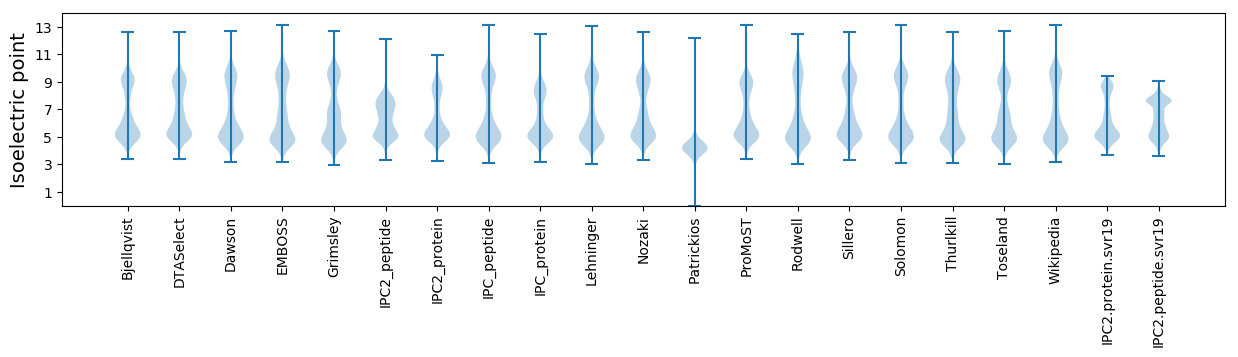
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L9MGR1|A0A3L9MGR1_9FLAO Uncharacterized protein OS=Flavobacteriaceae bacterium YIM 102668 OX=2479857 GN=EAH69_08200 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84KK12 pKa = 8.86KK13 pKa = 10.03RR14 pKa = 11.84NKK16 pKa = 9.59HH17 pKa = 4.05GFRR20 pKa = 11.84EE21 pKa = 4.13RR22 pKa = 11.84MASKK26 pKa = 10.63NGRR29 pKa = 11.84KK30 pKa = 9.21LLAARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.53KK38 pKa = 10.12GRR40 pKa = 11.84KK41 pKa = 8.55ALTVSSVRR49 pKa = 11.84TKK51 pKa = 10.73RR52 pKa = 3.29
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84KK12 pKa = 8.86KK13 pKa = 10.03RR14 pKa = 11.84NKK16 pKa = 9.59HH17 pKa = 4.05GFRR20 pKa = 11.84EE21 pKa = 4.13RR22 pKa = 11.84MASKK26 pKa = 10.63NGRR29 pKa = 11.84KK30 pKa = 9.21LLAARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.53KK38 pKa = 10.12GRR40 pKa = 11.84KK41 pKa = 8.55ALTVSSVRR49 pKa = 11.84TKK51 pKa = 10.73RR52 pKa = 3.29
Molecular weight: 6.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
857452 |
32 |
2398 |
321.5 |
36.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.976 ± 0.051 | 0.667 ± 0.014 |
5.596 ± 0.033 | 6.922 ± 0.049 |
5.426 ± 0.039 | 6.07 ± 0.044 |
1.642 ± 0.02 | 8.566 ± 0.052 |
8.026 ± 0.045 | 8.995 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.183 ± 0.022 | 6.808 ± 0.063 |
3.163 ± 0.025 | 3.663 ± 0.029 |
3.206 ± 0.033 | 6.142 ± 0.038 |
5.525 ± 0.04 | 6.141 ± 0.039 |
0.963 ± 0.017 | 4.32 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
