
Collinsella sp. An307
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; unclassified Collinsella
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
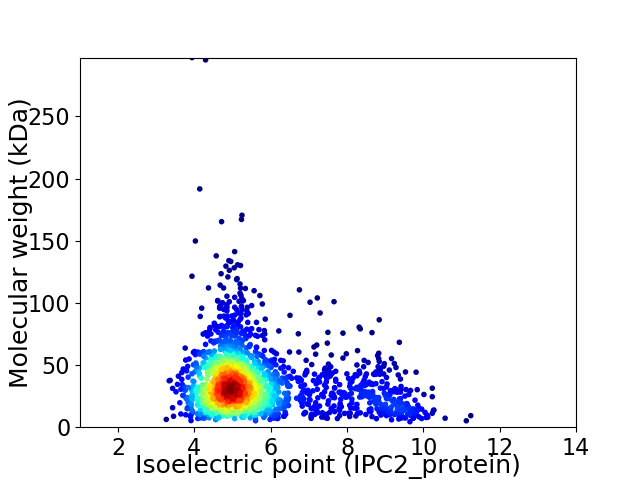
Virtual 2D-PAGE plot for 1893 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4AJA6|A0A1Y4AJA6_9ACTN Single-stranded-DNA-specific exonuclease RecJ OS=Collinsella sp. An307 OX=1965630 GN=B5F89_00375 PE=3 SV=1
MM1 pKa = 7.82KK2 pKa = 10.25DD3 pKa = 3.3LLVSRR8 pKa = 11.84RR9 pKa = 11.84SIAGFGVAAAAVALTACGNTAAEE32 pKa = 4.82GSAPAAGSASAEE44 pKa = 4.16SSDD47 pKa = 4.53GSVQLQVFAANSLEE61 pKa = 3.91KK62 pKa = 10.88AMPEE66 pKa = 3.88VEE68 pKa = 4.4ALYY71 pKa = 10.19TEE73 pKa = 4.24QTGVTFADD81 pKa = 3.64TQYY84 pKa = 11.05KK85 pKa = 10.56GSGDD89 pKa = 3.46LVEE92 pKa = 4.46QMRR95 pKa = 11.84GGAPVDD101 pKa = 3.36ILITASAGTMDD112 pKa = 4.44DD113 pKa = 3.73AEE115 pKa = 4.28EE116 pKa = 4.57AGLVDD121 pKa = 4.04ADD123 pKa = 3.64TRR125 pKa = 11.84EE126 pKa = 4.01DD127 pKa = 3.58MFVNDD132 pKa = 3.84LVIVRR137 pKa = 11.84AEE139 pKa = 4.05GSDD142 pKa = 3.5IEE144 pKa = 4.34ISSLKK149 pKa = 10.68DD150 pKa = 3.2VASVDD155 pKa = 3.47GNIAIGEE162 pKa = 4.22PGAVPAGKK170 pKa = 9.9YY171 pKa = 10.32ANQALASVGLYY182 pKa = 9.85TEE184 pKa = 5.27DD185 pKa = 3.29EE186 pKa = 4.8GEE188 pKa = 4.22GGEE191 pKa = 4.19YY192 pKa = 10.27DD193 pKa = 4.53ASIADD198 pKa = 4.03KK199 pKa = 10.83VVQADD204 pKa = 3.75KK205 pKa = 11.36VGTAAQYY212 pKa = 11.29VSTGDD217 pKa = 3.68CVIGFVYY224 pKa = 10.44TSDD227 pKa = 3.09VYY229 pKa = 10.75RR230 pKa = 11.84YY231 pKa = 10.44DD232 pKa = 4.21GIEE235 pKa = 3.86VAYY238 pKa = 7.82TCPAEE243 pKa = 3.97SHH245 pKa = 6.2KK246 pKa = 11.19AIVYY250 pKa = 8.37PGAVATSSEE259 pKa = 4.24NADD262 pKa = 3.34AAADD266 pKa = 4.02FLAFCMDD273 pKa = 3.91NEE275 pKa = 4.33DD276 pKa = 5.44ALAIWAEE283 pKa = 4.15YY284 pKa = 10.49GFEE287 pKa = 3.98LAA289 pKa = 5.71
MM1 pKa = 7.82KK2 pKa = 10.25DD3 pKa = 3.3LLVSRR8 pKa = 11.84RR9 pKa = 11.84SIAGFGVAAAAVALTACGNTAAEE32 pKa = 4.82GSAPAAGSASAEE44 pKa = 4.16SSDD47 pKa = 4.53GSVQLQVFAANSLEE61 pKa = 3.91KK62 pKa = 10.88AMPEE66 pKa = 3.88VEE68 pKa = 4.4ALYY71 pKa = 10.19TEE73 pKa = 4.24QTGVTFADD81 pKa = 3.64TQYY84 pKa = 11.05KK85 pKa = 10.56GSGDD89 pKa = 3.46LVEE92 pKa = 4.46QMRR95 pKa = 11.84GGAPVDD101 pKa = 3.36ILITASAGTMDD112 pKa = 4.44DD113 pKa = 3.73AEE115 pKa = 4.28EE116 pKa = 4.57AGLVDD121 pKa = 4.04ADD123 pKa = 3.64TRR125 pKa = 11.84EE126 pKa = 4.01DD127 pKa = 3.58MFVNDD132 pKa = 3.84LVIVRR137 pKa = 11.84AEE139 pKa = 4.05GSDD142 pKa = 3.5IEE144 pKa = 4.34ISSLKK149 pKa = 10.68DD150 pKa = 3.2VASVDD155 pKa = 3.47GNIAIGEE162 pKa = 4.22PGAVPAGKK170 pKa = 9.9YY171 pKa = 10.32ANQALASVGLYY182 pKa = 9.85TEE184 pKa = 5.27DD185 pKa = 3.29EE186 pKa = 4.8GEE188 pKa = 4.22GGEE191 pKa = 4.19YY192 pKa = 10.27DD193 pKa = 4.53ASIADD198 pKa = 4.03KK199 pKa = 10.83VVQADD204 pKa = 3.75KK205 pKa = 11.36VGTAAQYY212 pKa = 11.29VSTGDD217 pKa = 3.68CVIGFVYY224 pKa = 10.44TSDD227 pKa = 3.09VYY229 pKa = 10.75RR230 pKa = 11.84YY231 pKa = 10.44DD232 pKa = 4.21GIEE235 pKa = 3.86VAYY238 pKa = 7.82TCPAEE243 pKa = 3.97SHH245 pKa = 6.2KK246 pKa = 11.19AIVYY250 pKa = 8.37PGAVATSSEE259 pKa = 4.24NADD262 pKa = 3.34AAADD266 pKa = 4.02FLAFCMDD273 pKa = 3.91NEE275 pKa = 4.33DD276 pKa = 5.44ALAIWAEE283 pKa = 4.15YY284 pKa = 10.49GFEE287 pKa = 3.98LAA289 pKa = 5.71
Molecular weight: 29.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4ABJ6|A0A1Y4ABJ6_9ACTN Glycerol-3-phosphate dehydrogenase [NAD(P)+] OS=Collinsella sp. An307 OX=1965630 GN=gpsA PE=3 SV=1
MM1 pKa = 7.92PPRR4 pKa = 11.84PSLPRR9 pKa = 11.84SAPSTRR15 pKa = 11.84RR16 pKa = 11.84SRR18 pKa = 11.84PSSSRR23 pKa = 11.84RR24 pKa = 11.84APSSRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PRR34 pKa = 11.84SLIRR38 pKa = 11.84RR39 pKa = 11.84PSLSLRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84FSLRR51 pKa = 11.84TCPPTTRR58 pKa = 11.84RR59 pKa = 11.84GASALLTSSLARR71 pKa = 11.84RR72 pKa = 11.84TSPTSSRR79 pKa = 11.84ASSTT83 pKa = 3.3
MM1 pKa = 7.92PPRR4 pKa = 11.84PSLPRR9 pKa = 11.84SAPSTRR15 pKa = 11.84RR16 pKa = 11.84SRR18 pKa = 11.84PSSSRR23 pKa = 11.84RR24 pKa = 11.84APSSRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PRR34 pKa = 11.84SLIRR38 pKa = 11.84RR39 pKa = 11.84PSLSLRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84FSLRR51 pKa = 11.84TCPPTTRR58 pKa = 11.84RR59 pKa = 11.84GASALLTSSLARR71 pKa = 11.84RR72 pKa = 11.84TSPTSSRR79 pKa = 11.84ASSTT83 pKa = 3.3
Molecular weight: 9.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640865 |
37 |
2761 |
338.5 |
36.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.378 ± 0.079 | 1.451 ± 0.022 |
6.281 ± 0.048 | 6.876 ± 0.06 |
3.588 ± 0.04 | 8.141 ± 0.049 |
1.987 ± 0.024 | 5.178 ± 0.043 |
3.12 ± 0.043 | 9.258 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.537 ± 0.03 | 2.666 ± 0.026 |
4.428 ± 0.034 | 2.636 ± 0.027 |
6.597 ± 0.062 | 5.479 ± 0.05 |
5.578 ± 0.046 | 7.975 ± 0.042 |
1.058 ± 0.021 | 2.788 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
