
Pepper enamovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Enamovirus; unclassified Enamovirus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
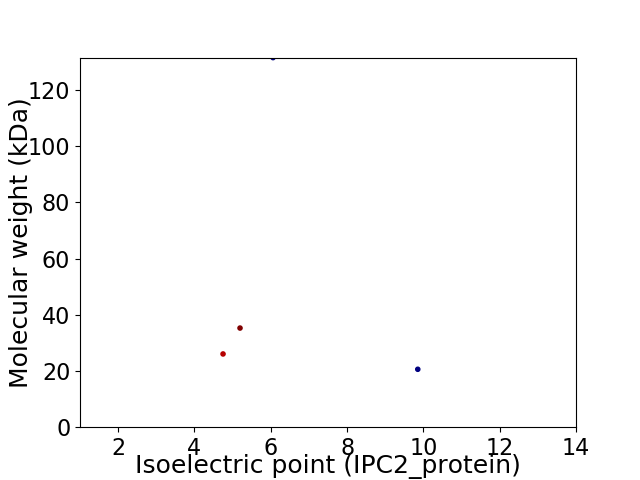
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
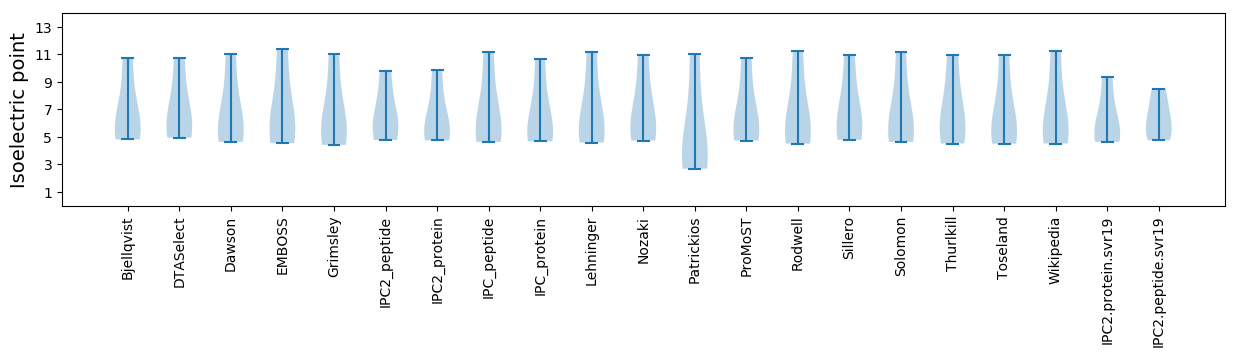
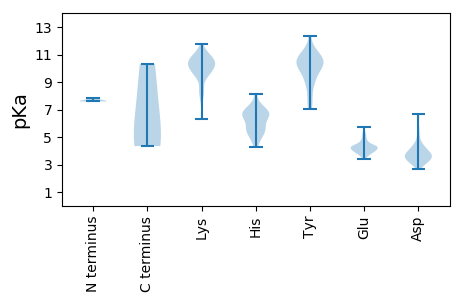
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I7UGC7|A0A2I7UGC7_9LUTE ORF0 OS=Pepper enamovirus OX=2073163 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.26ISTAEE7 pKa = 3.74NDD9 pKa = 3.18RR10 pKa = 11.84DD11 pKa = 3.56IVAKK15 pKa = 9.93SFNYY19 pKa = 9.84ARR21 pKa = 11.84FYY23 pKa = 10.82KK24 pKa = 10.26WEE26 pKa = 4.42DD27 pKa = 3.84DD28 pKa = 3.01KK29 pKa = 11.8WEE31 pKa = 4.24TVNLSAGFASNDD43 pKa = 3.48SEE45 pKa = 4.43KK46 pKa = 10.38AQPYY50 pKa = 10.04IIVPADD56 pKa = 3.0KK57 pKa = 11.17GKK59 pKa = 9.4FQVYY63 pKa = 8.99LEE65 pKa = 4.56CNGFQAVKK73 pKa = 10.5SIGGKK78 pKa = 9.98ADD80 pKa = 3.65GCWSGLVAYY89 pKa = 7.75EE90 pKa = 3.79ATGPGWLVNQYY101 pKa = 9.76VGCTITNYY109 pKa = 9.22QSSMKK114 pKa = 9.43FVCGHH119 pKa = 7.22PDD121 pKa = 3.18VEE123 pKa = 5.14LNGCKK128 pKa = 10.17FKK130 pKa = 11.26ANRR133 pKa = 11.84GVEE136 pKa = 3.78ADD138 pKa = 5.08FYY140 pKa = 11.66ASFDD144 pKa = 4.58LEE146 pKa = 5.76ADD148 pKa = 3.94DD149 pKa = 6.67DD150 pKa = 4.59DD151 pKa = 5.48SKK153 pKa = 11.27WVLYY157 pKa = 10.86APDD160 pKa = 3.74LRR162 pKa = 11.84KK163 pKa = 10.54DD164 pKa = 2.85SDD166 pKa = 3.77YY167 pKa = 11.3NYY169 pKa = 8.38VVSYY173 pKa = 10.9GGYY176 pKa = 7.21TDD178 pKa = 5.24KK179 pKa = 10.84ICEE182 pKa = 4.16VGSISISIDD191 pKa = 3.12EE192 pKa = 4.41VNAEE196 pKa = 4.08GSGSNAHH203 pKa = 5.66SALWRR208 pKa = 11.84LSEE211 pKa = 5.13PIDD214 pKa = 3.67KK215 pKa = 10.47EE216 pKa = 4.21KK217 pKa = 11.08SLKK220 pKa = 10.74LPLKK224 pKa = 10.43RR225 pKa = 11.84RR226 pKa = 11.84SCWSSDD232 pKa = 3.31SEE234 pKa = 4.35
MM1 pKa = 7.59KK2 pKa = 10.26ISTAEE7 pKa = 3.74NDD9 pKa = 3.18RR10 pKa = 11.84DD11 pKa = 3.56IVAKK15 pKa = 9.93SFNYY19 pKa = 9.84ARR21 pKa = 11.84FYY23 pKa = 10.82KK24 pKa = 10.26WEE26 pKa = 4.42DD27 pKa = 3.84DD28 pKa = 3.01KK29 pKa = 11.8WEE31 pKa = 4.24TVNLSAGFASNDD43 pKa = 3.48SEE45 pKa = 4.43KK46 pKa = 10.38AQPYY50 pKa = 10.04IIVPADD56 pKa = 3.0KK57 pKa = 11.17GKK59 pKa = 9.4FQVYY63 pKa = 8.99LEE65 pKa = 4.56CNGFQAVKK73 pKa = 10.5SIGGKK78 pKa = 9.98ADD80 pKa = 3.65GCWSGLVAYY89 pKa = 7.75EE90 pKa = 3.79ATGPGWLVNQYY101 pKa = 9.76VGCTITNYY109 pKa = 9.22QSSMKK114 pKa = 9.43FVCGHH119 pKa = 7.22PDD121 pKa = 3.18VEE123 pKa = 5.14LNGCKK128 pKa = 10.17FKK130 pKa = 11.26ANRR133 pKa = 11.84GVEE136 pKa = 3.78ADD138 pKa = 5.08FYY140 pKa = 11.66ASFDD144 pKa = 4.58LEE146 pKa = 5.76ADD148 pKa = 3.94DD149 pKa = 6.67DD150 pKa = 4.59DD151 pKa = 5.48SKK153 pKa = 11.27WVLYY157 pKa = 10.86APDD160 pKa = 3.74LRR162 pKa = 11.84KK163 pKa = 10.54DD164 pKa = 2.85SDD166 pKa = 3.77YY167 pKa = 11.3NYY169 pKa = 8.38VVSYY173 pKa = 10.9GGYY176 pKa = 7.21TDD178 pKa = 5.24KK179 pKa = 10.84ICEE182 pKa = 4.16VGSISISIDD191 pKa = 3.12EE192 pKa = 4.41VNAEE196 pKa = 4.08GSGSNAHH203 pKa = 5.66SALWRR208 pKa = 11.84LSEE211 pKa = 5.13PIDD214 pKa = 3.67KK215 pKa = 10.47EE216 pKa = 4.21KK217 pKa = 11.08SLKK220 pKa = 10.74LPLKK224 pKa = 10.43RR225 pKa = 11.84RR226 pKa = 11.84SCWSSDD232 pKa = 3.31SEE234 pKa = 4.35
Molecular weight: 26.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I7UGD2|A0A2I7UGD2_9LUTE Coat protein OS=Pepper enamovirus OX=2073163 PE=4 SV=1
MM1 pKa = 7.61ANNGRR6 pKa = 11.84RR7 pKa = 11.84NRR9 pKa = 11.84PRR11 pKa = 11.84RR12 pKa = 11.84AKK14 pKa = 9.82KK15 pKa = 10.02APRR18 pKa = 11.84AAPVVVVRR26 pKa = 11.84NAPARR31 pKa = 11.84PRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SGGGRR40 pKa = 11.84RR41 pKa = 11.84SRR43 pKa = 11.84RR44 pKa = 11.84GGGAGEE50 pKa = 3.67WRR52 pKa = 11.84PYY54 pKa = 10.36HH55 pKa = 7.27LYY57 pKa = 10.59GLKK60 pKa = 10.96GNDD63 pKa = 2.92KK64 pKa = 10.99GYY66 pKa = 8.98LTFGPDD72 pKa = 3.12GQTPSLGGGTLKK84 pKa = 10.82ACAEE88 pKa = 4.21YY89 pKa = 10.45KK90 pKa = 9.27ITALKK95 pKa = 9.49VQWKK99 pKa = 8.28SQASATVNGSMAIEE113 pKa = 4.67LGLGSTITTVSSRR126 pKa = 11.84ATSFKK131 pKa = 10.83LSVSGSKK138 pKa = 10.38SFSGKK143 pKa = 9.53EE144 pKa = 3.53LGATGRR150 pKa = 11.84MLPTGDD156 pKa = 3.67SASGEE161 pKa = 3.94KK162 pKa = 10.92GEE164 pKa = 4.17NQFRR168 pKa = 11.84LAYY171 pKa = 9.9NGNGPADD178 pKa = 3.37VCGDD182 pKa = 4.06LLCFFKK188 pKa = 10.78LQCAMPKK195 pKa = 10.31
MM1 pKa = 7.61ANNGRR6 pKa = 11.84RR7 pKa = 11.84NRR9 pKa = 11.84PRR11 pKa = 11.84RR12 pKa = 11.84AKK14 pKa = 9.82KK15 pKa = 10.02APRR18 pKa = 11.84AAPVVVVRR26 pKa = 11.84NAPARR31 pKa = 11.84PRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SGGGRR40 pKa = 11.84RR41 pKa = 11.84SRR43 pKa = 11.84RR44 pKa = 11.84GGGAGEE50 pKa = 3.67WRR52 pKa = 11.84PYY54 pKa = 10.36HH55 pKa = 7.27LYY57 pKa = 10.59GLKK60 pKa = 10.96GNDD63 pKa = 2.92KK64 pKa = 10.99GYY66 pKa = 8.98LTFGPDD72 pKa = 3.12GQTPSLGGGTLKK84 pKa = 10.82ACAEE88 pKa = 4.21YY89 pKa = 10.45KK90 pKa = 9.27ITALKK95 pKa = 9.49VQWKK99 pKa = 8.28SQASATVNGSMAIEE113 pKa = 4.67LGLGSTITTVSSRR126 pKa = 11.84ATSFKK131 pKa = 10.83LSVSGSKK138 pKa = 10.38SFSGKK143 pKa = 9.53EE144 pKa = 3.53LGATGRR150 pKa = 11.84MLPTGDD156 pKa = 3.67SASGEE161 pKa = 3.94KK162 pKa = 10.92GEE164 pKa = 4.17NQFRR168 pKa = 11.84LAYY171 pKa = 9.9NGNGPADD178 pKa = 3.37VCGDD182 pKa = 4.06LLCFFKK188 pKa = 10.78LQCAMPKK195 pKa = 10.31
Molecular weight: 20.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1936 |
195 |
1183 |
484.0 |
53.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.471 ± 0.316 | 2.634 ± 0.236 |
4.752 ± 0.993 | 6.87 ± 0.836 |
3.977 ± 0.233 | 8.419 ± 1.224 |
1.653 ± 0.468 | 3.254 ± 0.405 |
4.855 ± 1.096 | 10.176 ± 1.382 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.446 ± 0.219 | 2.996 ± 0.691 |
6.147 ± 0.886 | 3.461 ± 0.676 |
6.457 ± 0.942 | 8.523 ± 0.53 |
4.7 ± 0.5 | 6.302 ± 0.584 |
2.479 ± 0.356 | 2.428 ± 0.992 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
