
Allomyces macrogynus (strain ATCC 38327) (Allomyces javanicus var. macrogynus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Blastocladiomycota; Blastocladiomycota incertae sedis; Blastocladiomycetes; Blastocladiales; Blastocladiaceae; Allomyces; Allomyces macrogynus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
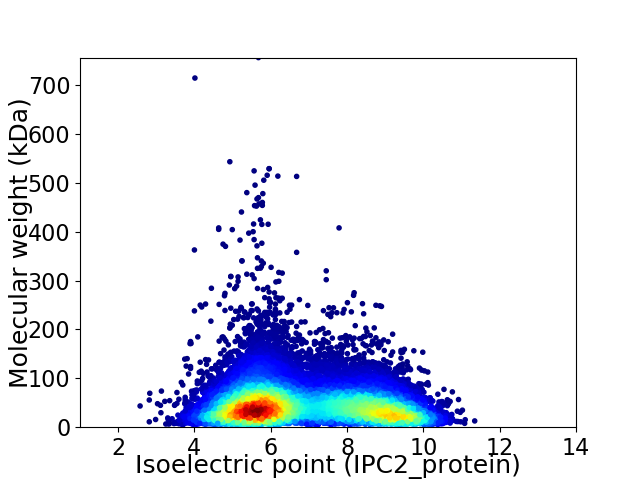
Virtual 2D-PAGE plot for 19092 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
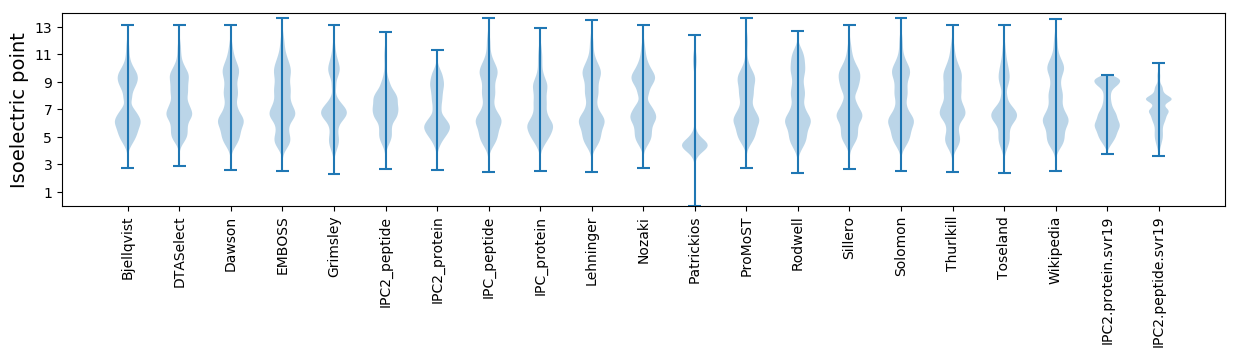
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L0SPN8|A0A0L0SPN8_ALLM3 Uncharacterized protein OS=Allomyces macrogynus (strain ATCC 38327) OX=578462 GN=AMAG_19162 PE=4 SV=1
MM1 pKa = 6.93PTAPRR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84PTRR12 pKa = 11.84ILAILATAALFAASFAHH29 pKa = 6.24ARR31 pKa = 11.84PVHH34 pKa = 6.4PYY36 pKa = 8.92GPPVAVPSRR45 pKa = 11.84APTSLDD51 pKa = 2.98AATPPMPVIVPPITSGSSTPDD72 pKa = 2.93VPVPVITRR80 pKa = 11.84PSGSTLSASTSVSSAMPTASEE101 pKa = 4.54SPRR104 pKa = 11.84SSVPSATTMASSAVYY119 pKa = 7.95TVPSASSTLVSSTSWPSTAIGTATTPIARR148 pKa = 11.84STAPSSVSTGGQVTATGTAIATSTAIITGSAPSPSARR185 pKa = 11.84TTTGTASATTSASATTSASATVSAPLSTNLSTASVGTTATTAYY228 pKa = 8.8GTDD231 pKa = 3.66TQSSSASSAIASTTAIAFTPYY252 pKa = 9.55PSSSTLPASEE262 pKa = 4.75SNGSEE267 pKa = 4.04LPTPTTTRR275 pKa = 11.84SADD278 pKa = 3.39VTTWPVDD285 pKa = 3.29PATTSLSTTSALPVPTVAPPASSDD309 pKa = 3.31STRR312 pKa = 11.84PTTGTYY318 pKa = 10.56VYY320 pKa = 10.39GAPTDD325 pKa = 3.93SASATPAPSTGTFVYY340 pKa = 10.56GSDD343 pKa = 3.45PTNAPVASPVSPGYY357 pKa = 10.45DD358 pKa = 2.76LGVGPTPTARR368 pKa = 11.84QDD370 pKa = 3.51PVASIPVATVTTIVVDD386 pKa = 3.92GEE388 pKa = 4.88VITSTVLMTPPADD401 pKa = 3.57ATPAPSPPAGGKK413 pKa = 9.23DD414 pKa = 3.49DD415 pKa = 5.44KK416 pKa = 11.15CDD418 pKa = 3.84CVCDD422 pKa = 4.96DD423 pKa = 5.53DD424 pKa = 5.61EE425 pKa = 4.96EE426 pKa = 4.43PANGDD431 pKa = 3.45DD432 pKa = 4.75CDD434 pKa = 4.07EE435 pKa = 4.59EE436 pKa = 5.13EE437 pKa = 4.36PAEE440 pKa = 4.52DD441 pKa = 4.28CDD443 pKa = 3.95EE444 pKa = 4.46EE445 pKa = 5.29EE446 pKa = 4.28PAEE449 pKa = 4.52DD450 pKa = 4.28CDD452 pKa = 3.95EE453 pKa = 4.46EE454 pKa = 5.29EE455 pKa = 4.52PAEE458 pKa = 4.48DD459 pKa = 4.5CDD461 pKa = 5.4DD462 pKa = 4.28EE463 pKa = 4.75EE464 pKa = 5.75EE465 pKa = 4.14EE466 pKa = 5.56AADD469 pKa = 4.82DD470 pKa = 4.51DD471 pKa = 4.55CDD473 pKa = 4.58EE474 pKa = 5.01EE475 pKa = 4.7GTTAAEE481 pKa = 4.3DD482 pKa = 3.84CTDD485 pKa = 3.39EE486 pKa = 4.31VVTVVVEE493 pKa = 3.94QDD495 pKa = 3.94CDD497 pKa = 3.9DD498 pKa = 5.07ADD500 pKa = 3.78PAAAKK505 pKa = 10.14GALEE509 pKa = 4.23VSTTAADD516 pKa = 3.42ATLTEE521 pKa = 4.61FAPVATTVAAEE532 pKa = 4.18TSSAASVITTSAPAAPASTSAAAPASTSSTAANTAAPAATPVVGTSEE579 pKa = 3.87NFYY582 pKa = 11.51
MM1 pKa = 6.93PTAPRR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84PTRR12 pKa = 11.84ILAILATAALFAASFAHH29 pKa = 6.24ARR31 pKa = 11.84PVHH34 pKa = 6.4PYY36 pKa = 8.92GPPVAVPSRR45 pKa = 11.84APTSLDD51 pKa = 2.98AATPPMPVIVPPITSGSSTPDD72 pKa = 2.93VPVPVITRR80 pKa = 11.84PSGSTLSASTSVSSAMPTASEE101 pKa = 4.54SPRR104 pKa = 11.84SSVPSATTMASSAVYY119 pKa = 7.95TVPSASSTLVSSTSWPSTAIGTATTPIARR148 pKa = 11.84STAPSSVSTGGQVTATGTAIATSTAIITGSAPSPSARR185 pKa = 11.84TTTGTASATTSASATTSASATVSAPLSTNLSTASVGTTATTAYY228 pKa = 8.8GTDD231 pKa = 3.66TQSSSASSAIASTTAIAFTPYY252 pKa = 9.55PSSSTLPASEE262 pKa = 4.75SNGSEE267 pKa = 4.04LPTPTTTRR275 pKa = 11.84SADD278 pKa = 3.39VTTWPVDD285 pKa = 3.29PATTSLSTTSALPVPTVAPPASSDD309 pKa = 3.31STRR312 pKa = 11.84PTTGTYY318 pKa = 10.56VYY320 pKa = 10.39GAPTDD325 pKa = 3.93SASATPAPSTGTFVYY340 pKa = 10.56GSDD343 pKa = 3.45PTNAPVASPVSPGYY357 pKa = 10.45DD358 pKa = 2.76LGVGPTPTARR368 pKa = 11.84QDD370 pKa = 3.51PVASIPVATVTTIVVDD386 pKa = 3.92GEE388 pKa = 4.88VITSTVLMTPPADD401 pKa = 3.57ATPAPSPPAGGKK413 pKa = 9.23DD414 pKa = 3.49DD415 pKa = 5.44KK416 pKa = 11.15CDD418 pKa = 3.84CVCDD422 pKa = 4.96DD423 pKa = 5.53DD424 pKa = 5.61EE425 pKa = 4.96EE426 pKa = 4.43PANGDD431 pKa = 3.45DD432 pKa = 4.75CDD434 pKa = 4.07EE435 pKa = 4.59EE436 pKa = 5.13EE437 pKa = 4.36PAEE440 pKa = 4.52DD441 pKa = 4.28CDD443 pKa = 3.95EE444 pKa = 4.46EE445 pKa = 5.29EE446 pKa = 4.28PAEE449 pKa = 4.52DD450 pKa = 4.28CDD452 pKa = 3.95EE453 pKa = 4.46EE454 pKa = 5.29EE455 pKa = 4.52PAEE458 pKa = 4.48DD459 pKa = 4.5CDD461 pKa = 5.4DD462 pKa = 4.28EE463 pKa = 4.75EE464 pKa = 5.75EE465 pKa = 4.14EE466 pKa = 5.56AADD469 pKa = 4.82DD470 pKa = 4.51DD471 pKa = 4.55CDD473 pKa = 4.58EE474 pKa = 5.01EE475 pKa = 4.7GTTAAEE481 pKa = 4.3DD482 pKa = 3.84CTDD485 pKa = 3.39EE486 pKa = 4.31VVTVVVEE493 pKa = 3.94QDD495 pKa = 3.94CDD497 pKa = 3.9DD498 pKa = 5.07ADD500 pKa = 3.78PAAAKK505 pKa = 10.14GALEE509 pKa = 4.23VSTTAADD516 pKa = 3.42ATLTEE521 pKa = 4.61FAPVATTVAAEE532 pKa = 4.18TSSAASVITTSAPAAPASTSAAAPASTSSTAANTAAPAATPVVGTSEE579 pKa = 3.87NFYY582 pKa = 11.51
Molecular weight: 57.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L0SEK3|A0A0L0SEK3_ALLM3 Uncharacterized protein OS=Allomyces macrogynus (strain ATCC 38327) OX=578462 GN=AMAG_06638 PE=4 SV=1
MM1 pKa = 7.75AGNPRR6 pKa = 11.84PTRR9 pKa = 11.84PTRR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84SPNKK19 pKa = 9.02RR20 pKa = 11.84FSKK23 pKa = 9.78ISSRR27 pKa = 11.84ISARR31 pKa = 11.84RR32 pKa = 11.84GSSRR36 pKa = 11.84RR37 pKa = 11.84SCAVPRR43 pKa = 11.84RR44 pKa = 11.84IRR46 pKa = 11.84WRR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84PRR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84AATRR61 pKa = 11.84RR62 pKa = 11.84GRR64 pKa = 11.84RR65 pKa = 11.84AGRR68 pKa = 11.84GRR70 pKa = 11.84RR71 pKa = 11.84TSCTTRR77 pKa = 11.84PRR79 pKa = 11.84NSRR82 pKa = 11.84RR83 pKa = 11.84VPRR86 pKa = 11.84RR87 pKa = 11.84ARR89 pKa = 11.84RR90 pKa = 11.84ARR92 pKa = 11.84IWAVPAGRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84AGRR106 pKa = 11.84GVTT109 pKa = 3.49
MM1 pKa = 7.75AGNPRR6 pKa = 11.84PTRR9 pKa = 11.84PTRR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84SPNKK19 pKa = 9.02RR20 pKa = 11.84FSKK23 pKa = 9.78ISSRR27 pKa = 11.84ISARR31 pKa = 11.84RR32 pKa = 11.84GSSRR36 pKa = 11.84RR37 pKa = 11.84SCAVPRR43 pKa = 11.84RR44 pKa = 11.84IRR46 pKa = 11.84WRR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84PRR54 pKa = 11.84RR55 pKa = 11.84ARR57 pKa = 11.84AATRR61 pKa = 11.84RR62 pKa = 11.84GRR64 pKa = 11.84RR65 pKa = 11.84AGRR68 pKa = 11.84GRR70 pKa = 11.84RR71 pKa = 11.84TSCTTRR77 pKa = 11.84PRR79 pKa = 11.84NSRR82 pKa = 11.84RR83 pKa = 11.84VPRR86 pKa = 11.84RR87 pKa = 11.84ARR89 pKa = 11.84RR90 pKa = 11.84ARR92 pKa = 11.84IWAVPAGRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84AGRR106 pKa = 11.84GVTT109 pKa = 3.49
Molecular weight: 12.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9242631 |
49 |
6944 |
484.1 |
52.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.312 ± 0.033 | 1.114 ± 0.006 |
5.842 ± 0.014 | 4.618 ± 0.016 |
2.992 ± 0.012 | 6.253 ± 0.015 |
2.743 ± 0.011 | 3.494 ± 0.013 |
3.534 ± 0.017 | 9.081 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.289 ± 0.007 | 2.58 ± 0.008 |
7.425 ± 0.027 | 3.588 ± 0.016 |
6.456 ± 0.016 | 7.215 ± 0.02 |
6.518 ± 0.015 | 7.565 ± 0.015 |
1.233 ± 0.006 | 2.147 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
