
Beihai weivirus-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.85
Get precalculated fractions of proteins
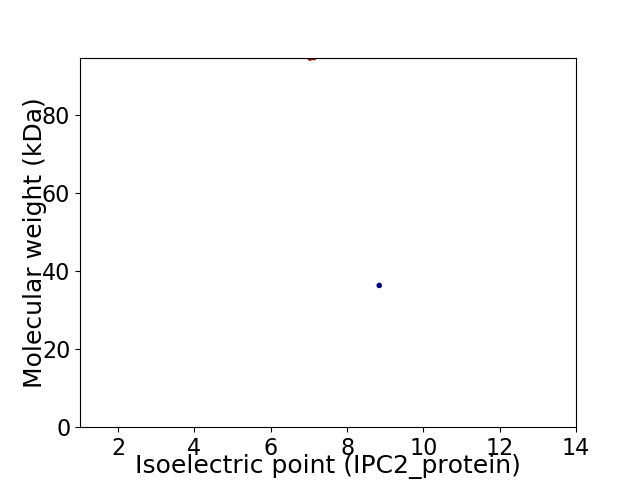
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL49|A0A1L3KL49_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 3 OX=1922751 PE=4 SV=1
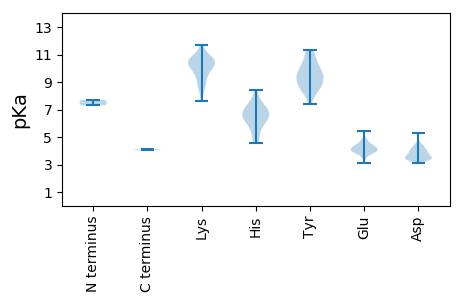
MM1 pKa = 7.31LASVPSGITWAYY13 pKa = 8.99DD14 pKa = 3.37APPEE18 pKa = 3.96LLEE21 pKa = 4.24IAGNRR26 pKa = 11.84AGSWKK31 pKa = 8.02ITTWEE36 pKa = 4.23FVHH39 pKa = 7.04PDD41 pKa = 3.13KK42 pKa = 11.27KK43 pKa = 10.58KK44 pKa = 10.63LKK46 pKa = 7.95STSVILTEE54 pKa = 5.18HH55 pKa = 6.69EE56 pKa = 4.61FNSLNATARR65 pKa = 11.84QCIASASKK73 pKa = 9.95EE74 pKa = 3.87QVLRR78 pKa = 11.84GSLEE82 pKa = 3.72QTAARR87 pKa = 11.84MWKK90 pKa = 8.5TGGLEE95 pKa = 4.01DD96 pKa = 3.81VALRR100 pKa = 11.84TLGPEE105 pKa = 4.06LVLRR109 pKa = 11.84WLRR112 pKa = 11.84AGQVQTGLTFHH123 pKa = 6.79FRR125 pKa = 11.84MGVPLSILSVLGACCCGSWRR145 pKa = 11.84TIVAILAKK153 pKa = 10.08KK154 pKa = 8.55VVKK157 pKa = 10.21FRR159 pKa = 11.84SIDD162 pKa = 3.43LRR164 pKa = 11.84LGLLVAAALLWDD176 pKa = 4.04AWRR179 pKa = 11.84SAGTYY184 pKa = 8.34RR185 pKa = 11.84TILRR189 pKa = 11.84WVGPSSSKK197 pKa = 9.39TSKK200 pKa = 9.83EE201 pKa = 3.86KK202 pKa = 10.89KK203 pKa = 7.62PAKK206 pKa = 9.82AAEE209 pKa = 4.41IKK211 pKa = 10.5DD212 pKa = 4.21DD213 pKa = 4.3PPKK216 pKa = 10.94ADD218 pKa = 4.55AGPSGDD224 pKa = 3.73KK225 pKa = 10.94VADD228 pKa = 3.8PSVSEE233 pKa = 4.0EE234 pKa = 3.88AEE236 pKa = 3.94ARR238 pKa = 11.84AKK240 pKa = 9.94PQEE243 pKa = 4.16NADD246 pKa = 3.67LAVRR250 pKa = 11.84TEE252 pKa = 4.01EE253 pKa = 3.82QEE255 pKa = 4.07ARR257 pKa = 11.84VTMEE261 pKa = 3.1GHH263 pKa = 4.54GRR265 pKa = 11.84YY266 pKa = 9.11RR267 pKa = 11.84IQMIEE272 pKa = 4.04DD273 pKa = 3.36KK274 pKa = 10.46KK275 pKa = 9.52VCVVLGQDD283 pKa = 3.49YY284 pKa = 10.87DD285 pKa = 4.08KK286 pKa = 11.33SVPKK290 pKa = 10.57DD291 pKa = 3.34RR292 pKa = 11.84FPDD295 pKa = 3.26VGVIVAPCSDD305 pKa = 4.29LPNVYY310 pKa = 9.5TNSAGNVHH318 pKa = 6.55HH319 pKa = 6.67GVEE322 pKa = 3.8EE323 pKa = 3.98RR324 pKa = 11.84LIKK327 pKa = 10.23KK328 pKa = 8.97SKK330 pKa = 8.77PCTWTKK336 pKa = 11.11DD337 pKa = 3.4DD338 pKa = 4.05KK339 pKa = 11.67LKK341 pKa = 10.78VGKK344 pKa = 10.08FIYY347 pKa = 10.28AAMGPKK353 pKa = 10.35GIFSTDD359 pKa = 3.17RR360 pKa = 11.84VRR362 pKa = 11.84SWYY365 pKa = 9.32EE366 pKa = 3.32SHH368 pKa = 7.5FALEE372 pKa = 4.73DD373 pKa = 3.42MRR375 pKa = 11.84SSKK378 pKa = 9.9WSEE381 pKa = 3.68SRR383 pKa = 11.84FVSTFEE389 pKa = 4.14GLLAQVNPTFKK400 pKa = 10.78FKK402 pKa = 9.98TAVKK406 pKa = 9.72AEE408 pKa = 4.04HH409 pKa = 6.12MPEE412 pKa = 3.85GKK414 pKa = 10.28APRR417 pKa = 11.84FLIADD422 pKa = 3.64GDD424 pKa = 4.08EE425 pKa = 4.49GQVLALAAVKK435 pKa = 10.51CMEE438 pKa = 4.09EE439 pKa = 3.91VLFEE443 pKa = 4.23VMEE446 pKa = 4.26SHH448 pKa = 7.44SIKK451 pKa = 10.47HH452 pKa = 4.58VCKK455 pKa = 10.17QEE457 pKa = 3.52AMKK460 pKa = 10.65RR461 pKa = 11.84LLGHH465 pKa = 6.1MVPPNAAKK473 pKa = 10.52KK474 pKa = 9.94AGCTFVEE481 pKa = 4.72GDD483 pKa = 3.28GSAWDD488 pKa = 4.34TTCSTAVRR496 pKa = 11.84GMVEE500 pKa = 4.11NPILQHH506 pKa = 6.25ISRR509 pKa = 11.84ILAQTYY515 pKa = 8.84IQPQSWAEE523 pKa = 3.84AHH525 pKa = 6.9DD526 pKa = 4.12KK527 pKa = 11.37ANAAQKK533 pKa = 10.64LKK535 pKa = 10.91LYY537 pKa = 9.49FKK539 pKa = 10.35KK540 pKa = 10.53YY541 pKa = 9.38HH542 pKa = 5.32EE543 pKa = 4.5TMHH546 pKa = 7.54VEE548 pKa = 3.72ITAIRR553 pKa = 11.84RR554 pKa = 11.84SGHH557 pKa = 6.32RR558 pKa = 11.84GTSVLNWWINFTMWVCSLFDD578 pKa = 3.49KK579 pKa = 10.76PQIFLCPDD587 pKa = 3.18ARR589 pKa = 11.84WADD592 pKa = 3.61DD593 pKa = 3.48VAGVRR598 pKa = 11.84RR599 pKa = 11.84WFYY602 pKa = 11.3GVYY605 pKa = 10.3EE606 pKa = 4.78GDD608 pKa = 4.01DD609 pKa = 4.03SGASTSPKK617 pKa = 9.3LCQVSEE623 pKa = 4.41ADD625 pKa = 3.45VTALKK630 pKa = 10.19EE631 pKa = 4.21KK632 pKa = 10.5KK633 pKa = 8.54ITLAEE638 pKa = 4.37LGNKK642 pKa = 8.8YY643 pKa = 9.66RR644 pKa = 11.84VPEE647 pKa = 4.1ASVTASISALAFWDD661 pKa = 3.58RR662 pKa = 11.84AGFNMKK668 pKa = 8.79WVFARR673 pKa = 11.84KK674 pKa = 8.87RR675 pKa = 11.84GTMVGCHH682 pKa = 6.57LGLTEE687 pKa = 4.52TDD689 pKa = 3.84SEE691 pKa = 4.89SKK693 pKa = 9.45TGCVVPNGVFCPEE706 pKa = 4.49LPRR709 pKa = 11.84ALKK712 pKa = 10.7GAVSCSPAIIEE723 pKa = 4.2AVRR726 pKa = 11.84KK727 pKa = 9.94GDD729 pKa = 3.46YY730 pKa = 9.08KK731 pKa = 10.33TIKK734 pKa = 9.41TIAAAAALGRR744 pKa = 11.84AADD747 pKa = 3.92FAGKK751 pKa = 10.4VPTLSRR757 pKa = 11.84KK758 pKa = 8.85YY759 pKa = 9.85LAYY762 pKa = 10.45ANEE765 pKa = 4.45LDD767 pKa = 4.17SSDD770 pKa = 3.94FEE772 pKa = 4.45DD773 pKa = 5.26RR774 pKa = 11.84EE775 pKa = 3.96MSMRR779 pKa = 11.84TVGEE783 pKa = 4.11EE784 pKa = 4.27GMSAAAVRR792 pKa = 11.84AQIEE796 pKa = 4.28EE797 pKa = 4.2ANGAVSAAEE806 pKa = 3.71EE807 pKa = 4.32KK808 pKa = 10.39TFLEE812 pKa = 4.27ALDD815 pKa = 3.77YY816 pKa = 8.46TASVEE821 pKa = 4.08EE822 pKa = 4.34LEE824 pKa = 4.51EE825 pKa = 4.01FCRR828 pKa = 11.84YY829 pKa = 8.39PWQFDD834 pKa = 3.34AVDD837 pKa = 3.4QHH839 pKa = 8.45DD840 pKa = 4.37EE841 pKa = 4.36FACSVPAAWRR851 pKa = 11.84DD852 pKa = 3.43GASII856 pKa = 4.04
MM1 pKa = 7.31LASVPSGITWAYY13 pKa = 8.99DD14 pKa = 3.37APPEE18 pKa = 3.96LLEE21 pKa = 4.24IAGNRR26 pKa = 11.84AGSWKK31 pKa = 8.02ITTWEE36 pKa = 4.23FVHH39 pKa = 7.04PDD41 pKa = 3.13KK42 pKa = 11.27KK43 pKa = 10.58KK44 pKa = 10.63LKK46 pKa = 7.95STSVILTEE54 pKa = 5.18HH55 pKa = 6.69EE56 pKa = 4.61FNSLNATARR65 pKa = 11.84QCIASASKK73 pKa = 9.95EE74 pKa = 3.87QVLRR78 pKa = 11.84GSLEE82 pKa = 3.72QTAARR87 pKa = 11.84MWKK90 pKa = 8.5TGGLEE95 pKa = 4.01DD96 pKa = 3.81VALRR100 pKa = 11.84TLGPEE105 pKa = 4.06LVLRR109 pKa = 11.84WLRR112 pKa = 11.84AGQVQTGLTFHH123 pKa = 6.79FRR125 pKa = 11.84MGVPLSILSVLGACCCGSWRR145 pKa = 11.84TIVAILAKK153 pKa = 10.08KK154 pKa = 8.55VVKK157 pKa = 10.21FRR159 pKa = 11.84SIDD162 pKa = 3.43LRR164 pKa = 11.84LGLLVAAALLWDD176 pKa = 4.04AWRR179 pKa = 11.84SAGTYY184 pKa = 8.34RR185 pKa = 11.84TILRR189 pKa = 11.84WVGPSSSKK197 pKa = 9.39TSKK200 pKa = 9.83EE201 pKa = 3.86KK202 pKa = 10.89KK203 pKa = 7.62PAKK206 pKa = 9.82AAEE209 pKa = 4.41IKK211 pKa = 10.5DD212 pKa = 4.21DD213 pKa = 4.3PPKK216 pKa = 10.94ADD218 pKa = 4.55AGPSGDD224 pKa = 3.73KK225 pKa = 10.94VADD228 pKa = 3.8PSVSEE233 pKa = 4.0EE234 pKa = 3.88AEE236 pKa = 3.94ARR238 pKa = 11.84AKK240 pKa = 9.94PQEE243 pKa = 4.16NADD246 pKa = 3.67LAVRR250 pKa = 11.84TEE252 pKa = 4.01EE253 pKa = 3.82QEE255 pKa = 4.07ARR257 pKa = 11.84VTMEE261 pKa = 3.1GHH263 pKa = 4.54GRR265 pKa = 11.84YY266 pKa = 9.11RR267 pKa = 11.84IQMIEE272 pKa = 4.04DD273 pKa = 3.36KK274 pKa = 10.46KK275 pKa = 9.52VCVVLGQDD283 pKa = 3.49YY284 pKa = 10.87DD285 pKa = 4.08KK286 pKa = 11.33SVPKK290 pKa = 10.57DD291 pKa = 3.34RR292 pKa = 11.84FPDD295 pKa = 3.26VGVIVAPCSDD305 pKa = 4.29LPNVYY310 pKa = 9.5TNSAGNVHH318 pKa = 6.55HH319 pKa = 6.67GVEE322 pKa = 3.8EE323 pKa = 3.98RR324 pKa = 11.84LIKK327 pKa = 10.23KK328 pKa = 8.97SKK330 pKa = 8.77PCTWTKK336 pKa = 11.11DD337 pKa = 3.4DD338 pKa = 4.05KK339 pKa = 11.67LKK341 pKa = 10.78VGKK344 pKa = 10.08FIYY347 pKa = 10.28AAMGPKK353 pKa = 10.35GIFSTDD359 pKa = 3.17RR360 pKa = 11.84VRR362 pKa = 11.84SWYY365 pKa = 9.32EE366 pKa = 3.32SHH368 pKa = 7.5FALEE372 pKa = 4.73DD373 pKa = 3.42MRR375 pKa = 11.84SSKK378 pKa = 9.9WSEE381 pKa = 3.68SRR383 pKa = 11.84FVSTFEE389 pKa = 4.14GLLAQVNPTFKK400 pKa = 10.78FKK402 pKa = 9.98TAVKK406 pKa = 9.72AEE408 pKa = 4.04HH409 pKa = 6.12MPEE412 pKa = 3.85GKK414 pKa = 10.28APRR417 pKa = 11.84FLIADD422 pKa = 3.64GDD424 pKa = 4.08EE425 pKa = 4.49GQVLALAAVKK435 pKa = 10.51CMEE438 pKa = 4.09EE439 pKa = 3.91VLFEE443 pKa = 4.23VMEE446 pKa = 4.26SHH448 pKa = 7.44SIKK451 pKa = 10.47HH452 pKa = 4.58VCKK455 pKa = 10.17QEE457 pKa = 3.52AMKK460 pKa = 10.65RR461 pKa = 11.84LLGHH465 pKa = 6.1MVPPNAAKK473 pKa = 10.52KK474 pKa = 9.94AGCTFVEE481 pKa = 4.72GDD483 pKa = 3.28GSAWDD488 pKa = 4.34TTCSTAVRR496 pKa = 11.84GMVEE500 pKa = 4.11NPILQHH506 pKa = 6.25ISRR509 pKa = 11.84ILAQTYY515 pKa = 8.84IQPQSWAEE523 pKa = 3.84AHH525 pKa = 6.9DD526 pKa = 4.12KK527 pKa = 11.37ANAAQKK533 pKa = 10.64LKK535 pKa = 10.91LYY537 pKa = 9.49FKK539 pKa = 10.35KK540 pKa = 10.53YY541 pKa = 9.38HH542 pKa = 5.32EE543 pKa = 4.5TMHH546 pKa = 7.54VEE548 pKa = 3.72ITAIRR553 pKa = 11.84RR554 pKa = 11.84SGHH557 pKa = 6.32RR558 pKa = 11.84GTSVLNWWINFTMWVCSLFDD578 pKa = 3.49KK579 pKa = 10.76PQIFLCPDD587 pKa = 3.18ARR589 pKa = 11.84WADD592 pKa = 3.61DD593 pKa = 3.48VAGVRR598 pKa = 11.84RR599 pKa = 11.84WFYY602 pKa = 11.3GVYY605 pKa = 10.3EE606 pKa = 4.78GDD608 pKa = 4.01DD609 pKa = 4.03SGASTSPKK617 pKa = 9.3LCQVSEE623 pKa = 4.41ADD625 pKa = 3.45VTALKK630 pKa = 10.19EE631 pKa = 4.21KK632 pKa = 10.5KK633 pKa = 8.54ITLAEE638 pKa = 4.37LGNKK642 pKa = 8.8YY643 pKa = 9.66RR644 pKa = 11.84VPEE647 pKa = 4.1ASVTASISALAFWDD661 pKa = 3.58RR662 pKa = 11.84AGFNMKK668 pKa = 8.79WVFARR673 pKa = 11.84KK674 pKa = 8.87RR675 pKa = 11.84GTMVGCHH682 pKa = 6.57LGLTEE687 pKa = 4.52TDD689 pKa = 3.84SEE691 pKa = 4.89SKK693 pKa = 9.45TGCVVPNGVFCPEE706 pKa = 4.49LPRR709 pKa = 11.84ALKK712 pKa = 10.7GAVSCSPAIIEE723 pKa = 4.2AVRR726 pKa = 11.84KK727 pKa = 9.94GDD729 pKa = 3.46YY730 pKa = 9.08KK731 pKa = 10.33TIKK734 pKa = 9.41TIAAAAALGRR744 pKa = 11.84AADD747 pKa = 3.92FAGKK751 pKa = 10.4VPTLSRR757 pKa = 11.84KK758 pKa = 8.85YY759 pKa = 9.85LAYY762 pKa = 10.45ANEE765 pKa = 4.45LDD767 pKa = 4.17SSDD770 pKa = 3.94FEE772 pKa = 4.45DD773 pKa = 5.26RR774 pKa = 11.84EE775 pKa = 3.96MSMRR779 pKa = 11.84TVGEE783 pKa = 4.11EE784 pKa = 4.27GMSAAAVRR792 pKa = 11.84AQIEE796 pKa = 4.28EE797 pKa = 4.2ANGAVSAAEE806 pKa = 3.71EE807 pKa = 4.32KK808 pKa = 10.39TFLEE812 pKa = 4.27ALDD815 pKa = 3.77YY816 pKa = 8.46TASVEE821 pKa = 4.08EE822 pKa = 4.34LEE824 pKa = 4.51EE825 pKa = 4.01FCRR828 pKa = 11.84YY829 pKa = 8.39PWQFDD834 pKa = 3.34AVDD837 pKa = 3.4QHH839 pKa = 8.45DD840 pKa = 4.37EE841 pKa = 4.36FACSVPAAWRR851 pKa = 11.84DD852 pKa = 3.43GASII856 pKa = 4.04
Molecular weight: 94.46 kDa
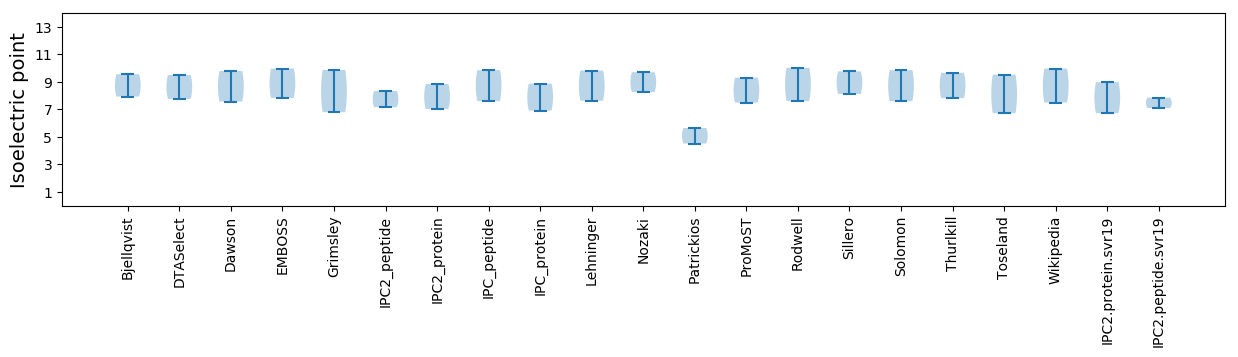
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL49|A0A1L3KL49_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 3 OX=1922751 PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 9.52TMRR6 pKa = 11.84AKK8 pKa = 10.5KK9 pKa = 9.8LARR12 pKa = 11.84DD13 pKa = 3.47LRR15 pKa = 11.84RR16 pKa = 11.84VKK18 pKa = 10.44RR19 pKa = 11.84DD20 pKa = 3.59LKK22 pKa = 10.88SKK24 pKa = 9.96PATRR28 pKa = 11.84RR29 pKa = 11.84NATKK33 pKa = 10.67ALAQAAMAAPRR44 pKa = 11.84RR45 pKa = 11.84NFGTKK50 pKa = 9.63SAGGTLRR57 pKa = 11.84ACIKK61 pKa = 10.72SLDD64 pKa = 3.57ARR66 pKa = 11.84VPRR69 pKa = 11.84TIGLPRR75 pKa = 11.84AVGPYY80 pKa = 9.95SVIRR84 pKa = 11.84TTRR87 pKa = 11.84LWSSSANFIMFVPFTSDD104 pKa = 3.79DD105 pKa = 3.54SWHH108 pKa = 5.74NWCGIEE114 pKa = 5.44DD115 pKa = 4.5VISSDD120 pKa = 3.97PVNGGNNSRR129 pKa = 11.84PITIPLADD137 pKa = 4.37LSHH140 pKa = 6.58AAEE143 pKa = 4.22VAPAALTVQVMNPSSVQTAQGMFAMGRR170 pKa = 11.84ANQQFDD176 pKa = 3.87YY177 pKa = 11.23GGSTEE182 pKa = 3.98TWDD185 pKa = 3.47TLRR188 pKa = 11.84DD189 pKa = 3.43RR190 pKa = 11.84FISYY194 pKa = 8.59FSPRR198 pKa = 11.84LLTGGKK204 pKa = 9.35LALRR208 pKa = 11.84GVKK211 pKa = 10.22CSTYY215 pKa = 9.6PLDD218 pKa = 3.46MSEE221 pKa = 4.78YY222 pKa = 8.54SHH224 pKa = 6.79FMPVADD230 pKa = 4.45YY231 pKa = 7.43PTPFTWLTVKK241 pKa = 9.1PTALAPIIFVQNNATPQTLEE261 pKa = 3.77FLITVEE267 pKa = 3.45WRR269 pKa = 11.84VRR271 pKa = 11.84FDD273 pKa = 3.48PGNPAVASHH282 pKa = 5.96THH284 pKa = 6.79HH285 pKa = 7.32DD286 pKa = 3.8TTPDD290 pKa = 3.69GVWNSVCRR298 pKa = 11.84EE299 pKa = 3.97MSSMGHH305 pKa = 5.21GVEE308 pKa = 4.48EE309 pKa = 4.02LAEE312 pKa = 4.14DD313 pKa = 3.98VMEE316 pKa = 4.79LGAMGGAARR325 pKa = 11.84LAATALLL332 pKa = 4.11
MM1 pKa = 7.69AKK3 pKa = 9.52TMRR6 pKa = 11.84AKK8 pKa = 10.5KK9 pKa = 9.8LARR12 pKa = 11.84DD13 pKa = 3.47LRR15 pKa = 11.84RR16 pKa = 11.84VKK18 pKa = 10.44RR19 pKa = 11.84DD20 pKa = 3.59LKK22 pKa = 10.88SKK24 pKa = 9.96PATRR28 pKa = 11.84RR29 pKa = 11.84NATKK33 pKa = 10.67ALAQAAMAAPRR44 pKa = 11.84RR45 pKa = 11.84NFGTKK50 pKa = 9.63SAGGTLRR57 pKa = 11.84ACIKK61 pKa = 10.72SLDD64 pKa = 3.57ARR66 pKa = 11.84VPRR69 pKa = 11.84TIGLPRR75 pKa = 11.84AVGPYY80 pKa = 9.95SVIRR84 pKa = 11.84TTRR87 pKa = 11.84LWSSSANFIMFVPFTSDD104 pKa = 3.79DD105 pKa = 3.54SWHH108 pKa = 5.74NWCGIEE114 pKa = 5.44DD115 pKa = 4.5VISSDD120 pKa = 3.97PVNGGNNSRR129 pKa = 11.84PITIPLADD137 pKa = 4.37LSHH140 pKa = 6.58AAEE143 pKa = 4.22VAPAALTVQVMNPSSVQTAQGMFAMGRR170 pKa = 11.84ANQQFDD176 pKa = 3.87YY177 pKa = 11.23GGSTEE182 pKa = 3.98TWDD185 pKa = 3.47TLRR188 pKa = 11.84DD189 pKa = 3.43RR190 pKa = 11.84FISYY194 pKa = 8.59FSPRR198 pKa = 11.84LLTGGKK204 pKa = 9.35LALRR208 pKa = 11.84GVKK211 pKa = 10.22CSTYY215 pKa = 9.6PLDD218 pKa = 3.46MSEE221 pKa = 4.78YY222 pKa = 8.54SHH224 pKa = 6.79FMPVADD230 pKa = 4.45YY231 pKa = 7.43PTPFTWLTVKK241 pKa = 9.1PTALAPIIFVQNNATPQTLEE261 pKa = 3.77FLITVEE267 pKa = 3.45WRR269 pKa = 11.84VRR271 pKa = 11.84FDD273 pKa = 3.48PGNPAVASHH282 pKa = 5.96THH284 pKa = 6.79HH285 pKa = 7.32DD286 pKa = 3.8TTPDD290 pKa = 3.69GVWNSVCRR298 pKa = 11.84EE299 pKa = 3.97MSSMGHH305 pKa = 5.21GVEE308 pKa = 4.48EE309 pKa = 4.02LAEE312 pKa = 4.14DD313 pKa = 3.98VMEE316 pKa = 4.79LGAMGGAARR325 pKa = 11.84LAATALLL332 pKa = 4.11
Molecular weight: 36.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1188 |
332 |
856 |
594.0 |
65.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.785 ± 0.02 | 2.02 ± 0.435 |
5.219 ± 0.052 | 6.229 ± 1.555 |
3.704 ± 0.113 | 6.566 ± 0.032 |
2.104 ± 0.002 | 3.956 ± 0.182 |
6.313 ± 1.439 | 7.492 ± 0.181 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.607 | 2.609 ± 0.696 |
4.966 ± 0.885 | 2.525 ± 0.062 |
6.145 ± 0.739 | 7.323 ± 0.11 |
6.313 ± 1.131 | 7.407 ± 0.416 |
2.525 ± 0.222 | 2.02 ± 0.114 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
