
Ustilaginoidea virens RNA virus M
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 5.58
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

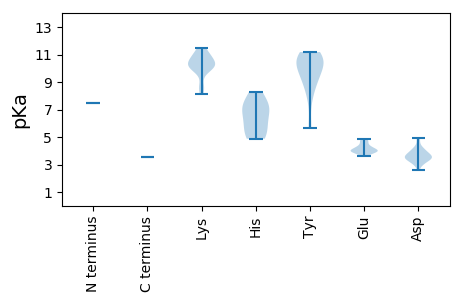
Protein with the lowest isoelectric point:
>tr|A0A097I5B9|A0A097I5B9_9VIRU RNA-directed RNA polymerase OS=Ustilaginoidea virens RNA virus M OX=1561172 PE=4 SV=1
MM1 pKa = 7.47SEE3 pKa = 4.27YY4 pKa = 10.56VPPSIEE10 pKa = 4.74DD11 pKa = 3.34LWKK14 pKa = 10.12HH15 pKa = 5.04LQGFGANPIALRR27 pKa = 11.84DD28 pKa = 4.18HH29 pKa = 6.0ITVPVTEE36 pKa = 4.62MLSEE40 pKa = 4.5LRR42 pKa = 11.84GLDD45 pKa = 3.38LGRR48 pKa = 11.84GRR50 pKa = 11.84IAGWLEE56 pKa = 3.62PEE58 pKa = 4.25NILRR62 pKa = 11.84VEE64 pKa = 4.51VPAATSPGIRR74 pKa = 11.84WKK76 pKa = 10.87KK77 pKa = 10.21LGYY80 pKa = 7.89KK81 pKa = 8.13TKK83 pKa = 10.39RR84 pKa = 11.84DD85 pKa = 3.57AMLQAAVEE93 pKa = 3.96ARR95 pKa = 11.84GNIVRR100 pKa = 11.84MRR102 pKa = 11.84DD103 pKa = 3.05VGGGYY108 pKa = 8.53EE109 pKa = 4.06VPPCGVAGRR118 pKa = 11.84GKK120 pKa = 10.03RR121 pKa = 11.84VPIDD125 pKa = 3.41RR126 pKa = 11.84PSDD129 pKa = 3.62GEE131 pKa = 4.0RR132 pKa = 11.84KK133 pKa = 8.69EE134 pKa = 3.95GRR136 pKa = 11.84FIVMPDD142 pKa = 3.48LVRR145 pKa = 11.84HH146 pKa = 5.6LMGMIAVQPLMKK158 pKa = 10.1KK159 pKa = 10.09IKK161 pKa = 10.22QADD164 pKa = 3.56KK165 pKa = 11.3SNGGVMLGMGPFGGSYY181 pKa = 10.83DD182 pKa = 3.66RR183 pKa = 11.84LAEE186 pKa = 4.01YY187 pKa = 10.84ASGGKK192 pKa = 10.05AYY194 pKa = 10.86LCIDD198 pKa = 4.32FKK200 pKa = 11.46KK201 pKa = 10.48FDD203 pKa = 3.52QSVPRR208 pKa = 11.84SVLRR212 pKa = 11.84AVMQRR217 pKa = 11.84VRR219 pKa = 11.84EE220 pKa = 4.05EE221 pKa = 3.77FDD223 pKa = 3.38EE224 pKa = 4.82EE225 pKa = 4.26EE226 pKa = 4.81GSDD229 pKa = 4.05GYY231 pKa = 10.05WRR233 pKa = 11.84SEE235 pKa = 3.8FKK237 pKa = 10.88QLVDD241 pKa = 4.03TEE243 pKa = 4.31VAMPDD248 pKa = 2.59GHH250 pKa = 6.59TYY252 pKa = 10.81AKK254 pKa = 10.18RR255 pKa = 11.84AGVSSGDD262 pKa = 3.16PWTSIVGSVANWLVLRR278 pKa = 11.84IALKK282 pKa = 10.64RR283 pKa = 11.84LGVDD287 pKa = 3.08ARR289 pKa = 11.84IWTFGDD295 pKa = 3.46DD296 pKa = 3.39SLIAVDD302 pKa = 3.82TDD304 pKa = 3.77VPSEE308 pKa = 4.07SWLLEE313 pKa = 3.71AVARR317 pKa = 11.84EE318 pKa = 4.09LKK320 pKa = 10.53SVFGMTVSKK329 pKa = 10.08EE330 pKa = 3.6KK331 pKa = 10.84SIATRR336 pKa = 11.84HH337 pKa = 4.88LVGVFEE343 pKa = 4.35EE344 pKa = 4.87PEE346 pKa = 4.02VGQTANFLSAYY357 pKa = 9.41FLQTGTSVVPVRR369 pKa = 11.84PIQDD373 pKa = 3.45LHH375 pKa = 8.27EE376 pKa = 4.67LMLVPEE382 pKa = 4.22RR383 pKa = 11.84RR384 pKa = 11.84KK385 pKa = 9.72DD386 pKa = 3.78TVWWEE391 pKa = 4.0VARR394 pKa = 11.84TAMAYY399 pKa = 10.38LVFFWNDD406 pKa = 3.02SARR409 pKa = 11.84MTLEE413 pKa = 3.89WYY415 pKa = 9.27WDD417 pKa = 3.77FLHH420 pKa = 7.19RR421 pKa = 11.84EE422 pKa = 3.94YY423 pKa = 10.64EE424 pKa = 4.24IPQLRR429 pKa = 11.84GDD431 pKa = 3.94PEE433 pKa = 3.95ILGMLRR439 pKa = 11.84LMDD442 pKa = 4.95LPWDD446 pKa = 3.64QFKK449 pKa = 11.25EE450 pKa = 3.89EE451 pKa = 3.91WLIRR455 pKa = 11.84LPYY458 pKa = 9.68RR459 pKa = 11.84YY460 pKa = 9.16EE461 pKa = 3.85VEE463 pKa = 4.03LLYY466 pKa = 11.18SHH468 pKa = 7.32GYY470 pKa = 5.68TTFFPPALWALTYY483 pKa = 10.21KK484 pKa = 10.84DD485 pKa = 4.76HH486 pKa = 7.28VPLDD490 pKa = 4.11GGNKK494 pKa = 8.64VWWEE498 pKa = 4.0LEE500 pKa = 4.14GQQ502 pKa = 3.54
MM1 pKa = 7.47SEE3 pKa = 4.27YY4 pKa = 10.56VPPSIEE10 pKa = 4.74DD11 pKa = 3.34LWKK14 pKa = 10.12HH15 pKa = 5.04LQGFGANPIALRR27 pKa = 11.84DD28 pKa = 4.18HH29 pKa = 6.0ITVPVTEE36 pKa = 4.62MLSEE40 pKa = 4.5LRR42 pKa = 11.84GLDD45 pKa = 3.38LGRR48 pKa = 11.84GRR50 pKa = 11.84IAGWLEE56 pKa = 3.62PEE58 pKa = 4.25NILRR62 pKa = 11.84VEE64 pKa = 4.51VPAATSPGIRR74 pKa = 11.84WKK76 pKa = 10.87KK77 pKa = 10.21LGYY80 pKa = 7.89KK81 pKa = 8.13TKK83 pKa = 10.39RR84 pKa = 11.84DD85 pKa = 3.57AMLQAAVEE93 pKa = 3.96ARR95 pKa = 11.84GNIVRR100 pKa = 11.84MRR102 pKa = 11.84DD103 pKa = 3.05VGGGYY108 pKa = 8.53EE109 pKa = 4.06VPPCGVAGRR118 pKa = 11.84GKK120 pKa = 10.03RR121 pKa = 11.84VPIDD125 pKa = 3.41RR126 pKa = 11.84PSDD129 pKa = 3.62GEE131 pKa = 4.0RR132 pKa = 11.84KK133 pKa = 8.69EE134 pKa = 3.95GRR136 pKa = 11.84FIVMPDD142 pKa = 3.48LVRR145 pKa = 11.84HH146 pKa = 5.6LMGMIAVQPLMKK158 pKa = 10.1KK159 pKa = 10.09IKK161 pKa = 10.22QADD164 pKa = 3.56KK165 pKa = 11.3SNGGVMLGMGPFGGSYY181 pKa = 10.83DD182 pKa = 3.66RR183 pKa = 11.84LAEE186 pKa = 4.01YY187 pKa = 10.84ASGGKK192 pKa = 10.05AYY194 pKa = 10.86LCIDD198 pKa = 4.32FKK200 pKa = 11.46KK201 pKa = 10.48FDD203 pKa = 3.52QSVPRR208 pKa = 11.84SVLRR212 pKa = 11.84AVMQRR217 pKa = 11.84VRR219 pKa = 11.84EE220 pKa = 4.05EE221 pKa = 3.77FDD223 pKa = 3.38EE224 pKa = 4.82EE225 pKa = 4.26EE226 pKa = 4.81GSDD229 pKa = 4.05GYY231 pKa = 10.05WRR233 pKa = 11.84SEE235 pKa = 3.8FKK237 pKa = 10.88QLVDD241 pKa = 4.03TEE243 pKa = 4.31VAMPDD248 pKa = 2.59GHH250 pKa = 6.59TYY252 pKa = 10.81AKK254 pKa = 10.18RR255 pKa = 11.84AGVSSGDD262 pKa = 3.16PWTSIVGSVANWLVLRR278 pKa = 11.84IALKK282 pKa = 10.64RR283 pKa = 11.84LGVDD287 pKa = 3.08ARR289 pKa = 11.84IWTFGDD295 pKa = 3.46DD296 pKa = 3.39SLIAVDD302 pKa = 3.82TDD304 pKa = 3.77VPSEE308 pKa = 4.07SWLLEE313 pKa = 3.71AVARR317 pKa = 11.84EE318 pKa = 4.09LKK320 pKa = 10.53SVFGMTVSKK329 pKa = 10.08EE330 pKa = 3.6KK331 pKa = 10.84SIATRR336 pKa = 11.84HH337 pKa = 4.88LVGVFEE343 pKa = 4.35EE344 pKa = 4.87PEE346 pKa = 4.02VGQTANFLSAYY357 pKa = 9.41FLQTGTSVVPVRR369 pKa = 11.84PIQDD373 pKa = 3.45LHH375 pKa = 8.27EE376 pKa = 4.67LMLVPEE382 pKa = 4.22RR383 pKa = 11.84RR384 pKa = 11.84KK385 pKa = 9.72DD386 pKa = 3.78TVWWEE391 pKa = 4.0VARR394 pKa = 11.84TAMAYY399 pKa = 10.38LVFFWNDD406 pKa = 3.02SARR409 pKa = 11.84MTLEE413 pKa = 3.89WYY415 pKa = 9.27WDD417 pKa = 3.77FLHH420 pKa = 7.19RR421 pKa = 11.84EE422 pKa = 3.94YY423 pKa = 10.64EE424 pKa = 4.24IPQLRR429 pKa = 11.84GDD431 pKa = 3.94PEE433 pKa = 3.95ILGMLRR439 pKa = 11.84LMDD442 pKa = 4.95LPWDD446 pKa = 3.64QFKK449 pKa = 11.25EE450 pKa = 3.89EE451 pKa = 3.91WLIRR455 pKa = 11.84LPYY458 pKa = 9.68RR459 pKa = 11.84YY460 pKa = 9.16EE461 pKa = 3.85VEE463 pKa = 4.03LLYY466 pKa = 11.18SHH468 pKa = 7.32GYY470 pKa = 5.68TTFFPPALWALTYY483 pKa = 10.21KK484 pKa = 10.84DD485 pKa = 4.76HH486 pKa = 7.28VPLDD490 pKa = 4.11GGNKK494 pKa = 8.64VWWEE498 pKa = 4.0LEE500 pKa = 4.14GQQ502 pKa = 3.54
Molecular weight: 57.1 kDa
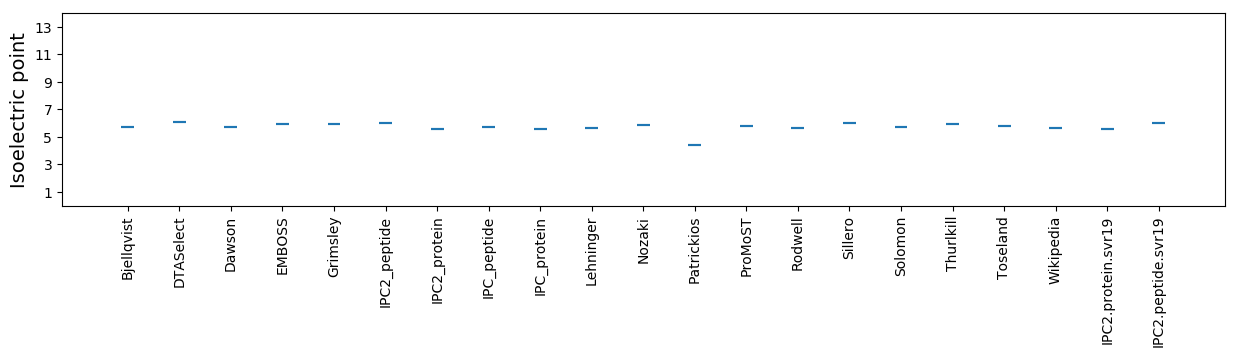
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097I5B9|A0A097I5B9_9VIRU RNA-directed RNA polymerase OS=Ustilaginoidea virens RNA virus M OX=1561172 PE=4 SV=1
MM1 pKa = 7.47SEE3 pKa = 4.27YY4 pKa = 10.56VPPSIEE10 pKa = 4.74DD11 pKa = 3.34LWKK14 pKa = 10.12HH15 pKa = 5.04LQGFGANPIALRR27 pKa = 11.84DD28 pKa = 4.18HH29 pKa = 6.0ITVPVTEE36 pKa = 4.62MLSEE40 pKa = 4.5LRR42 pKa = 11.84GLDD45 pKa = 3.38LGRR48 pKa = 11.84GRR50 pKa = 11.84IAGWLEE56 pKa = 3.62PEE58 pKa = 4.25NILRR62 pKa = 11.84VEE64 pKa = 4.51VPAATSPGIRR74 pKa = 11.84WKK76 pKa = 10.87KK77 pKa = 10.21LGYY80 pKa = 7.89KK81 pKa = 8.13TKK83 pKa = 10.39RR84 pKa = 11.84DD85 pKa = 3.57AMLQAAVEE93 pKa = 3.96ARR95 pKa = 11.84GNIVRR100 pKa = 11.84MRR102 pKa = 11.84DD103 pKa = 3.05VGGGYY108 pKa = 8.53EE109 pKa = 4.06VPPCGVAGRR118 pKa = 11.84GKK120 pKa = 10.03RR121 pKa = 11.84VPIDD125 pKa = 3.41RR126 pKa = 11.84PSDD129 pKa = 3.62GEE131 pKa = 4.0RR132 pKa = 11.84KK133 pKa = 8.69EE134 pKa = 3.95GRR136 pKa = 11.84FIVMPDD142 pKa = 3.48LVRR145 pKa = 11.84HH146 pKa = 5.6LMGMIAVQPLMKK158 pKa = 10.1KK159 pKa = 10.09IKK161 pKa = 10.22QADD164 pKa = 3.56KK165 pKa = 11.3SNGGVMLGMGPFGGSYY181 pKa = 10.83DD182 pKa = 3.66RR183 pKa = 11.84LAEE186 pKa = 4.01YY187 pKa = 10.84ASGGKK192 pKa = 10.05AYY194 pKa = 10.86LCIDD198 pKa = 4.32FKK200 pKa = 11.46KK201 pKa = 10.48FDD203 pKa = 3.52QSVPRR208 pKa = 11.84SVLRR212 pKa = 11.84AVMQRR217 pKa = 11.84VRR219 pKa = 11.84EE220 pKa = 4.05EE221 pKa = 3.77FDD223 pKa = 3.38EE224 pKa = 4.82EE225 pKa = 4.26EE226 pKa = 4.81GSDD229 pKa = 4.05GYY231 pKa = 10.05WRR233 pKa = 11.84SEE235 pKa = 3.8FKK237 pKa = 10.88QLVDD241 pKa = 4.03TEE243 pKa = 4.31VAMPDD248 pKa = 2.59GHH250 pKa = 6.59TYY252 pKa = 10.81AKK254 pKa = 10.18RR255 pKa = 11.84AGVSSGDD262 pKa = 3.16PWTSIVGSVANWLVLRR278 pKa = 11.84IALKK282 pKa = 10.64RR283 pKa = 11.84LGVDD287 pKa = 3.08ARR289 pKa = 11.84IWTFGDD295 pKa = 3.46DD296 pKa = 3.39SLIAVDD302 pKa = 3.82TDD304 pKa = 3.77VPSEE308 pKa = 4.07SWLLEE313 pKa = 3.71AVARR317 pKa = 11.84EE318 pKa = 4.09LKK320 pKa = 10.53SVFGMTVSKK329 pKa = 10.08EE330 pKa = 3.6KK331 pKa = 10.84SIATRR336 pKa = 11.84HH337 pKa = 4.88LVGVFEE343 pKa = 4.35EE344 pKa = 4.87PEE346 pKa = 4.02VGQTANFLSAYY357 pKa = 9.41FLQTGTSVVPVRR369 pKa = 11.84PIQDD373 pKa = 3.45LHH375 pKa = 8.27EE376 pKa = 4.67LMLVPEE382 pKa = 4.22RR383 pKa = 11.84RR384 pKa = 11.84KK385 pKa = 9.72DD386 pKa = 3.78TVWWEE391 pKa = 4.0VARR394 pKa = 11.84TAMAYY399 pKa = 10.38LVFFWNDD406 pKa = 3.02SARR409 pKa = 11.84MTLEE413 pKa = 3.89WYY415 pKa = 9.27WDD417 pKa = 3.77FLHH420 pKa = 7.19RR421 pKa = 11.84EE422 pKa = 3.94YY423 pKa = 10.64EE424 pKa = 4.24IPQLRR429 pKa = 11.84GDD431 pKa = 3.94PEE433 pKa = 3.95ILGMLRR439 pKa = 11.84LMDD442 pKa = 4.95LPWDD446 pKa = 3.64QFKK449 pKa = 11.25EE450 pKa = 3.89EE451 pKa = 3.91WLIRR455 pKa = 11.84LPYY458 pKa = 9.68RR459 pKa = 11.84YY460 pKa = 9.16EE461 pKa = 3.85VEE463 pKa = 4.03LLYY466 pKa = 11.18SHH468 pKa = 7.32GYY470 pKa = 5.68TTFFPPALWALTYY483 pKa = 10.21KK484 pKa = 10.84DD485 pKa = 4.76HH486 pKa = 7.28VPLDD490 pKa = 4.11GGNKK494 pKa = 8.64VWWEE498 pKa = 4.0LEE500 pKa = 4.14GQQ502 pKa = 3.54
MM1 pKa = 7.47SEE3 pKa = 4.27YY4 pKa = 10.56VPPSIEE10 pKa = 4.74DD11 pKa = 3.34LWKK14 pKa = 10.12HH15 pKa = 5.04LQGFGANPIALRR27 pKa = 11.84DD28 pKa = 4.18HH29 pKa = 6.0ITVPVTEE36 pKa = 4.62MLSEE40 pKa = 4.5LRR42 pKa = 11.84GLDD45 pKa = 3.38LGRR48 pKa = 11.84GRR50 pKa = 11.84IAGWLEE56 pKa = 3.62PEE58 pKa = 4.25NILRR62 pKa = 11.84VEE64 pKa = 4.51VPAATSPGIRR74 pKa = 11.84WKK76 pKa = 10.87KK77 pKa = 10.21LGYY80 pKa = 7.89KK81 pKa = 8.13TKK83 pKa = 10.39RR84 pKa = 11.84DD85 pKa = 3.57AMLQAAVEE93 pKa = 3.96ARR95 pKa = 11.84GNIVRR100 pKa = 11.84MRR102 pKa = 11.84DD103 pKa = 3.05VGGGYY108 pKa = 8.53EE109 pKa = 4.06VPPCGVAGRR118 pKa = 11.84GKK120 pKa = 10.03RR121 pKa = 11.84VPIDD125 pKa = 3.41RR126 pKa = 11.84PSDD129 pKa = 3.62GEE131 pKa = 4.0RR132 pKa = 11.84KK133 pKa = 8.69EE134 pKa = 3.95GRR136 pKa = 11.84FIVMPDD142 pKa = 3.48LVRR145 pKa = 11.84HH146 pKa = 5.6LMGMIAVQPLMKK158 pKa = 10.1KK159 pKa = 10.09IKK161 pKa = 10.22QADD164 pKa = 3.56KK165 pKa = 11.3SNGGVMLGMGPFGGSYY181 pKa = 10.83DD182 pKa = 3.66RR183 pKa = 11.84LAEE186 pKa = 4.01YY187 pKa = 10.84ASGGKK192 pKa = 10.05AYY194 pKa = 10.86LCIDD198 pKa = 4.32FKK200 pKa = 11.46KK201 pKa = 10.48FDD203 pKa = 3.52QSVPRR208 pKa = 11.84SVLRR212 pKa = 11.84AVMQRR217 pKa = 11.84VRR219 pKa = 11.84EE220 pKa = 4.05EE221 pKa = 3.77FDD223 pKa = 3.38EE224 pKa = 4.82EE225 pKa = 4.26EE226 pKa = 4.81GSDD229 pKa = 4.05GYY231 pKa = 10.05WRR233 pKa = 11.84SEE235 pKa = 3.8FKK237 pKa = 10.88QLVDD241 pKa = 4.03TEE243 pKa = 4.31VAMPDD248 pKa = 2.59GHH250 pKa = 6.59TYY252 pKa = 10.81AKK254 pKa = 10.18RR255 pKa = 11.84AGVSSGDD262 pKa = 3.16PWTSIVGSVANWLVLRR278 pKa = 11.84IALKK282 pKa = 10.64RR283 pKa = 11.84LGVDD287 pKa = 3.08ARR289 pKa = 11.84IWTFGDD295 pKa = 3.46DD296 pKa = 3.39SLIAVDD302 pKa = 3.82TDD304 pKa = 3.77VPSEE308 pKa = 4.07SWLLEE313 pKa = 3.71AVARR317 pKa = 11.84EE318 pKa = 4.09LKK320 pKa = 10.53SVFGMTVSKK329 pKa = 10.08EE330 pKa = 3.6KK331 pKa = 10.84SIATRR336 pKa = 11.84HH337 pKa = 4.88LVGVFEE343 pKa = 4.35EE344 pKa = 4.87PEE346 pKa = 4.02VGQTANFLSAYY357 pKa = 9.41FLQTGTSVVPVRR369 pKa = 11.84PIQDD373 pKa = 3.45LHH375 pKa = 8.27EE376 pKa = 4.67LMLVPEE382 pKa = 4.22RR383 pKa = 11.84RR384 pKa = 11.84KK385 pKa = 9.72DD386 pKa = 3.78TVWWEE391 pKa = 4.0VARR394 pKa = 11.84TAMAYY399 pKa = 10.38LVFFWNDD406 pKa = 3.02SARR409 pKa = 11.84MTLEE413 pKa = 3.89WYY415 pKa = 9.27WDD417 pKa = 3.77FLHH420 pKa = 7.19RR421 pKa = 11.84EE422 pKa = 3.94YY423 pKa = 10.64EE424 pKa = 4.24IPQLRR429 pKa = 11.84GDD431 pKa = 3.94PEE433 pKa = 3.95ILGMLRR439 pKa = 11.84LMDD442 pKa = 4.95LPWDD446 pKa = 3.64QFKK449 pKa = 11.25EE450 pKa = 3.89EE451 pKa = 3.91WLIRR455 pKa = 11.84LPYY458 pKa = 9.68RR459 pKa = 11.84YY460 pKa = 9.16EE461 pKa = 3.85VEE463 pKa = 4.03LLYY466 pKa = 11.18SHH468 pKa = 7.32GYY470 pKa = 5.68TTFFPPALWALTYY483 pKa = 10.21KK484 pKa = 10.84DD485 pKa = 4.76HH486 pKa = 7.28VPLDD490 pKa = 4.11GGNKK494 pKa = 8.64VWWEE498 pKa = 4.0LEE500 pKa = 4.14GQQ502 pKa = 3.54
Molecular weight: 57.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
502 |
502 |
502 |
502.0 |
57.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.773 ± 0.0 | 0.398 ± 0.0 |
6.175 ± 0.0 | 7.769 ± 0.0 |
3.586 ± 0.0 | 8.765 ± 0.0 |
1.793 ± 0.0 | 4.183 ± 0.0 |
4.781 ± 0.0 | 9.761 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.586 ± 0.0 | 1.594 ± 0.0 |
5.777 ± 0.0 | 2.59 ± 0.0 |
7.57 ± 0.0 | 5.179 ± 0.0 |
3.984 ± 0.0 | 8.765 ± 0.0 |
3.586 ± 0.0 | 3.386 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
