
Candidatus Marinamargulisbacteria bacterium SCGC AG-333-B06
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Candidatus Margulisbacteria; Candidatus Marinamargulisbacteria
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
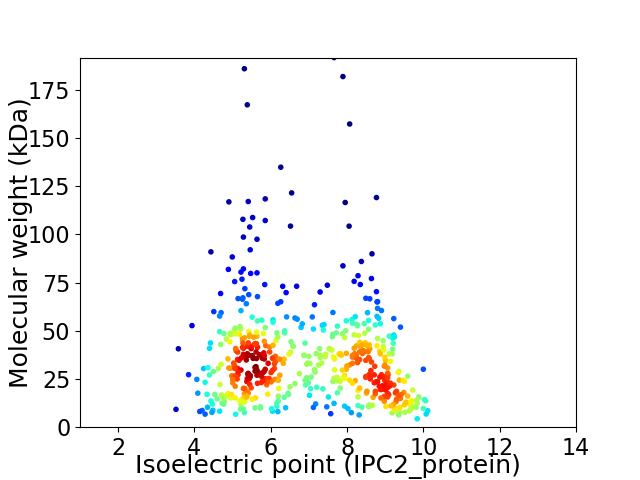
Virtual 2D-PAGE plot for 544 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328R783|A0A328R783_9BACT NTP_transferase domain-containing protein OS=Candidatus Marinamargulisbacteria bacterium SCGC AG-333-B06 OX=2184342 GN=DID74_01640 PE=4 SV=1
MM1 pKa = 7.83PKK3 pKa = 9.89HH4 pKa = 6.53LFRR7 pKa = 11.84ILCACFLSLSLLLTSCGNLLDD28 pKa = 5.05DD29 pKa = 5.34ASSSDD34 pKa = 3.37SDD36 pKa = 3.69TSRR39 pKa = 11.84GALSITDD46 pKa = 3.57ITMDD50 pKa = 3.89SYY52 pKa = 11.17PVSFMEE58 pKa = 5.29EE59 pKa = 3.38IQKK62 pKa = 10.79NIEE65 pKa = 3.88ASAGEE70 pKa = 4.64LFTAASYY77 pKa = 11.04SVNQYY82 pKa = 10.18NLVYY86 pKa = 9.27HH87 pKa = 6.92TIDD90 pKa = 3.02AKK92 pKa = 11.42GNATLASAKK101 pKa = 10.56LLIPDD106 pKa = 4.78ANTDD110 pKa = 3.49LDD112 pKa = 4.28LVVYY116 pKa = 9.09LHH118 pKa = 6.52GTTSLDD124 pKa = 3.12ADD126 pKa = 4.09APTNAYY132 pKa = 10.65ADD134 pKa = 3.84FSDD137 pKa = 4.98FNGDD141 pKa = 3.42DD142 pKa = 3.48ATTIDD147 pKa = 5.01HH148 pKa = 7.05NIADD152 pKa = 4.07MKK154 pKa = 8.29EE155 pKa = 3.99TAMLMSFVSDD165 pKa = 3.27TNIAVLMPDD174 pKa = 3.37YY175 pKa = 10.97LGFGASEE182 pKa = 4.09GLHH185 pKa = 6.56PYY187 pKa = 8.4MHH189 pKa = 6.93GASLASATIDD199 pKa = 4.14AIRR202 pKa = 11.84AALNFASLSSNPFSLTGDD220 pKa = 3.82FTLTGYY226 pKa = 10.36SEE228 pKa = 4.05GGYY231 pKa = 11.02AVMATHH237 pKa = 7.54KK238 pKa = 10.54EE239 pKa = 3.86LLADD243 pKa = 3.74NGTVEE248 pKa = 4.57SAYY251 pKa = 10.8LDD253 pKa = 3.3IMGSEE258 pKa = 4.57GARR261 pKa = 11.84LKK263 pKa = 10.95AVIPGAPPCNISQRR277 pKa = 11.84MAAVMLPPAGIAYY290 pKa = 7.78PAPAFAAYY298 pKa = 9.55VALAYY303 pKa = 10.84NEE305 pKa = 4.41IYY307 pKa = 10.46DD308 pKa = 3.69IEE310 pKa = 5.04ANISTIFSPTYY321 pKa = 10.57NYY323 pKa = 10.17IIDD326 pKa = 4.88DD327 pKa = 3.85FDD329 pKa = 4.2GSKK332 pKa = 10.79SLTTMTGEE340 pKa = 4.27FLAGGIFPPTPISMFSTEE358 pKa = 4.09FQDD361 pKa = 6.05AILADD366 pKa = 3.65YY367 pKa = 9.44TKK369 pKa = 10.46ILEE372 pKa = 5.05DD373 pKa = 3.61PTATPDD379 pKa = 3.38DD380 pKa = 4.22DD381 pKa = 5.47FYY383 pKa = 11.96QKK385 pKa = 10.69FKK387 pKa = 11.18EE388 pKa = 3.92NDD390 pKa = 3.2VFDD393 pKa = 4.18YY394 pKa = 11.17HH395 pKa = 7.13PGTAYY400 pKa = 11.07YY401 pKa = 9.76EE402 pKa = 4.39IIYY405 pKa = 8.04TASDD409 pKa = 3.9EE410 pKa = 4.27IVPSINATEE419 pKa = 3.96AAEE422 pKa = 4.01ALAVPGRR429 pKa = 11.84SVNILFDD436 pKa = 3.92PAEE439 pKa = 4.31ISSDD443 pKa = 3.25PTGINLFADD452 pKa = 4.54PQDD455 pKa = 3.63STYY458 pKa = 9.89VTIKK462 pKa = 10.87AGDD465 pKa = 3.79PSLLIHH471 pKa = 6.77SSAAGFYY478 pKa = 9.92IGRR481 pKa = 11.84SFNVSKK487 pKa = 10.77MMLSQQ492 pKa = 3.5
MM1 pKa = 7.83PKK3 pKa = 9.89HH4 pKa = 6.53LFRR7 pKa = 11.84ILCACFLSLSLLLTSCGNLLDD28 pKa = 5.05DD29 pKa = 5.34ASSSDD34 pKa = 3.37SDD36 pKa = 3.69TSRR39 pKa = 11.84GALSITDD46 pKa = 3.57ITMDD50 pKa = 3.89SYY52 pKa = 11.17PVSFMEE58 pKa = 5.29EE59 pKa = 3.38IQKK62 pKa = 10.79NIEE65 pKa = 3.88ASAGEE70 pKa = 4.64LFTAASYY77 pKa = 11.04SVNQYY82 pKa = 10.18NLVYY86 pKa = 9.27HH87 pKa = 6.92TIDD90 pKa = 3.02AKK92 pKa = 11.42GNATLASAKK101 pKa = 10.56LLIPDD106 pKa = 4.78ANTDD110 pKa = 3.49LDD112 pKa = 4.28LVVYY116 pKa = 9.09LHH118 pKa = 6.52GTTSLDD124 pKa = 3.12ADD126 pKa = 4.09APTNAYY132 pKa = 10.65ADD134 pKa = 3.84FSDD137 pKa = 4.98FNGDD141 pKa = 3.42DD142 pKa = 3.48ATTIDD147 pKa = 5.01HH148 pKa = 7.05NIADD152 pKa = 4.07MKK154 pKa = 8.29EE155 pKa = 3.99TAMLMSFVSDD165 pKa = 3.27TNIAVLMPDD174 pKa = 3.37YY175 pKa = 10.97LGFGASEE182 pKa = 4.09GLHH185 pKa = 6.56PYY187 pKa = 8.4MHH189 pKa = 6.93GASLASATIDD199 pKa = 4.14AIRR202 pKa = 11.84AALNFASLSSNPFSLTGDD220 pKa = 3.82FTLTGYY226 pKa = 10.36SEE228 pKa = 4.05GGYY231 pKa = 11.02AVMATHH237 pKa = 7.54KK238 pKa = 10.54EE239 pKa = 3.86LLADD243 pKa = 3.74NGTVEE248 pKa = 4.57SAYY251 pKa = 10.8LDD253 pKa = 3.3IMGSEE258 pKa = 4.57GARR261 pKa = 11.84LKK263 pKa = 10.95AVIPGAPPCNISQRR277 pKa = 11.84MAAVMLPPAGIAYY290 pKa = 7.78PAPAFAAYY298 pKa = 9.55VALAYY303 pKa = 10.84NEE305 pKa = 4.41IYY307 pKa = 10.46DD308 pKa = 3.69IEE310 pKa = 5.04ANISTIFSPTYY321 pKa = 10.57NYY323 pKa = 10.17IIDD326 pKa = 4.88DD327 pKa = 3.85FDD329 pKa = 4.2GSKK332 pKa = 10.79SLTTMTGEE340 pKa = 4.27FLAGGIFPPTPISMFSTEE358 pKa = 4.09FQDD361 pKa = 6.05AILADD366 pKa = 3.65YY367 pKa = 9.44TKK369 pKa = 10.46ILEE372 pKa = 5.05DD373 pKa = 3.61PTATPDD379 pKa = 3.38DD380 pKa = 4.22DD381 pKa = 5.47FYY383 pKa = 11.96QKK385 pKa = 10.69FKK387 pKa = 11.18EE388 pKa = 3.92NDD390 pKa = 3.2VFDD393 pKa = 4.18YY394 pKa = 11.17HH395 pKa = 7.13PGTAYY400 pKa = 11.07YY401 pKa = 9.76EE402 pKa = 4.39IIYY405 pKa = 8.04TASDD409 pKa = 3.9EE410 pKa = 4.27IVPSINATEE419 pKa = 3.96AAEE422 pKa = 4.01ALAVPGRR429 pKa = 11.84SVNILFDD436 pKa = 3.92PAEE439 pKa = 4.31ISSDD443 pKa = 3.25PTGINLFADD452 pKa = 4.54PQDD455 pKa = 3.63STYY458 pKa = 9.89VTIKK462 pKa = 10.87AGDD465 pKa = 3.79PSLLIHH471 pKa = 6.77SSAAGFYY478 pKa = 9.92IGRR481 pKa = 11.84SFNVSKK487 pKa = 10.77MMLSQQ492 pKa = 3.5
Molecular weight: 52.68 kDa
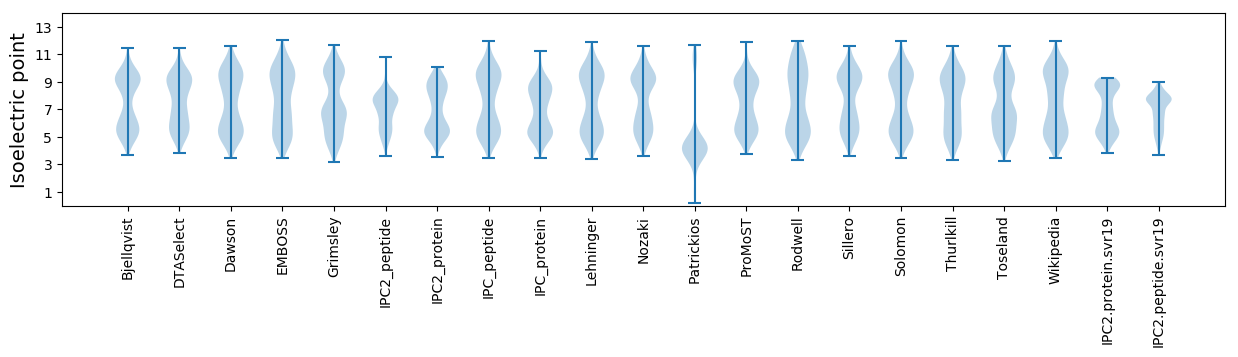
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328QZQ4|A0A328QZQ4_9BACT AB hydrolase-1 domain-containing protein OS=Candidatus Marinamargulisbacteria bacterium SCGC AG-333-B06 OX=2184342 GN=DID74_01160 PE=4 SV=1
MM1 pKa = 6.54YY2 pKa = 8.73TYY4 pKa = 10.97FSMQSSIISKK14 pKa = 10.77ANLAKK19 pKa = 10.31EE20 pKa = 4.44LAVHH24 pKa = 6.45MGWTQKK30 pKa = 10.88KK31 pKa = 9.5SLLFIDD37 pKa = 5.88DD38 pKa = 3.73CFDD41 pKa = 4.04WIKK44 pKa = 10.93SQMLRR49 pKa = 11.84GHH51 pKa = 7.17RR52 pKa = 11.84IVLSGFGVFQLNLRR66 pKa = 11.84KK67 pKa = 9.5KK68 pKa = 10.26RR69 pKa = 11.84RR70 pKa = 11.84LLHH73 pKa = 6.79PMTQKK78 pKa = 10.69PLLVPEE84 pKa = 4.35RR85 pKa = 11.84LRR87 pKa = 11.84PNFTPSDD94 pKa = 3.81AVLHH98 pKa = 5.9LLNHH102 pKa = 6.87RR103 pKa = 11.84ARR105 pKa = 11.84LL106 pKa = 3.66
MM1 pKa = 6.54YY2 pKa = 8.73TYY4 pKa = 10.97FSMQSSIISKK14 pKa = 10.77ANLAKK19 pKa = 10.31EE20 pKa = 4.44LAVHH24 pKa = 6.45MGWTQKK30 pKa = 10.88KK31 pKa = 9.5SLLFIDD37 pKa = 5.88DD38 pKa = 3.73CFDD41 pKa = 4.04WIKK44 pKa = 10.93SQMLRR49 pKa = 11.84GHH51 pKa = 7.17RR52 pKa = 11.84IVLSGFGVFQLNLRR66 pKa = 11.84KK67 pKa = 9.5KK68 pKa = 10.26RR69 pKa = 11.84RR70 pKa = 11.84LLHH73 pKa = 6.79PMTQKK78 pKa = 10.69PLLVPEE84 pKa = 4.35RR85 pKa = 11.84LRR87 pKa = 11.84PNFTPSDD94 pKa = 3.81AVLHH98 pKa = 5.9LLNHH102 pKa = 6.87RR103 pKa = 11.84ARR105 pKa = 11.84LL106 pKa = 3.66
Molecular weight: 12.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
177059 |
37 |
1667 |
325.5 |
36.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.551 ± 0.109 | 1.296 ± 0.044 |
5.601 ± 0.096 | 5.317 ± 0.111 |
5.085 ± 0.106 | 5.495 ± 0.105 |
2.504 ± 0.061 | 9.116 ± 0.122 |
7.673 ± 0.135 | 10.176 ± 0.123 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.176 ± 0.051 | 5.618 ± 0.098 |
3.578 ± 0.062 | 3.852 ± 0.064 |
3.324 ± 0.081 | 7.587 ± 0.099 |
5.603 ± 0.077 | 5.554 ± 0.084 |
0.842 ± 0.033 | 4.053 ± 0.072 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
