
Sanxia water strider virus 13
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.33
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
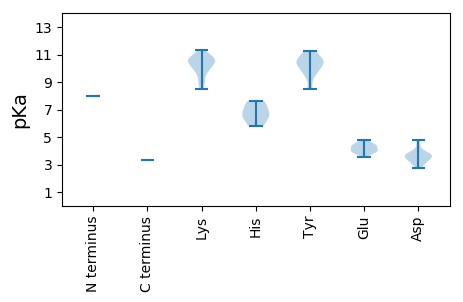
Protein with the lowest isoelectric point:
>tr|A0A1L3KIK1|A0A1L3KIK1_9VIRU RNA-dependent RNA polymerase OS=Sanxia water strider virus 13 OX=1923397 PE=4 SV=1
MM1 pKa = 7.99PLQSMPSQKK10 pKa = 10.66VNFGSLSAVAASLRR24 pKa = 11.84RR25 pKa = 11.84LARR28 pKa = 11.84EE29 pKa = 3.56GRR31 pKa = 11.84TVGSSQYY38 pKa = 10.36ATFARR43 pKa = 11.84LAEE46 pKa = 4.3KK47 pKa = 10.7LRR49 pKa = 11.84GLSPRR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84CLSTASWVAGGLSVSTTDD76 pKa = 3.2RR77 pKa = 11.84SKK79 pKa = 11.35GSSLGHH85 pKa = 6.33RR86 pKa = 11.84LAVVKK91 pKa = 9.18FAIKK95 pKa = 10.01LWNNIISDD103 pKa = 3.89VEE105 pKa = 4.25QFVVDD110 pKa = 3.87YY111 pKa = 11.28KK112 pKa = 10.7RR113 pKa = 11.84VCNSAAYY120 pKa = 9.64AAVRR124 pKa = 11.84GYY126 pKa = 9.62TSIVAAGKK134 pKa = 10.27CRR136 pKa = 11.84DD137 pKa = 3.62YY138 pKa = 11.27LHH140 pKa = 7.24RR141 pKa = 11.84AFGPCGIIPHH151 pKa = 6.69KK152 pKa = 10.52GRR154 pKa = 11.84RR155 pKa = 11.84SLFALASTTRR165 pKa = 11.84CLPPASLTPSRR176 pKa = 11.84VKK178 pKa = 10.49IEE180 pKa = 4.0LEE182 pKa = 3.84ALKK185 pKa = 10.83SRR187 pKa = 11.84LTSVPPPVSGALLSEE202 pKa = 4.36LVSFSHH208 pKa = 7.63DD209 pKa = 3.69LLRR212 pKa = 11.84DD213 pKa = 3.69AGPPVQVEE221 pKa = 4.32VTGNALGPPGKK232 pKa = 9.05TSLDD236 pKa = 3.28RR237 pKa = 11.84AQLYY241 pKa = 10.44GSYY244 pKa = 10.42AFKK247 pKa = 10.77GVIMSKK253 pKa = 8.76TLLMMSKK260 pKa = 10.11SAIPPDD266 pKa = 3.69VPFKK270 pKa = 11.03EE271 pKa = 4.33DD272 pKa = 3.47LEE274 pKa = 4.75EE275 pKa = 4.03NVQRR279 pKa = 11.84VSLLAEE285 pKa = 4.65AGDD288 pKa = 3.83KK289 pKa = 10.57VRR291 pKa = 11.84AITVPSSLAVQLAGRR306 pKa = 11.84SINRR310 pKa = 11.84ALIRR314 pKa = 11.84CLRR317 pKa = 11.84RR318 pKa = 11.84SDD320 pKa = 3.46VFGPGFGKK328 pKa = 10.66RR329 pKa = 11.84PGTLMSGANLGDD341 pKa = 3.56KK342 pKa = 10.68HH343 pKa = 7.46IVSCDD348 pKa = 3.02LTAASDD354 pKa = 3.94YY355 pKa = 11.12LNQDD359 pKa = 2.83VAIAVLADD367 pKa = 3.47YY368 pKa = 10.59LRR370 pKa = 11.84SHH372 pKa = 7.36ASGIWYY378 pKa = 8.48PWASALLGPQRR389 pKa = 11.84IRR391 pKa = 11.84SRR393 pKa = 11.84DD394 pKa = 3.41DD395 pKa = 3.11TYY397 pKa = 8.57TTSRR401 pKa = 11.84GVLMGTPLAWPILSICHH418 pKa = 6.79AFACYY423 pKa = 9.55KK424 pKa = 10.81ALGNTYY430 pKa = 10.43QFLCRR435 pKa = 11.84IVGDD439 pKa = 4.64DD440 pKa = 4.77GLLICNASQYY450 pKa = 9.63RR451 pKa = 11.84AYY453 pKa = 10.61VSVMNRR459 pKa = 11.84LGFVINEE466 pKa = 4.06HH467 pKa = 6.19KK468 pKa = 9.68TFQGKK473 pKa = 8.52QCGILAGRR481 pKa = 11.84LYY483 pKa = 10.81VLKK486 pKa = 9.86TRR488 pKa = 11.84RR489 pKa = 11.84AKK491 pKa = 10.62FQDD494 pKa = 3.58LNSPTKK500 pKa = 8.93TMAWRR505 pKa = 11.84MVDD508 pKa = 3.32CQAPYY513 pKa = 10.74VPEE516 pKa = 4.05LLAARR521 pKa = 11.84GAQRR525 pKa = 11.84FGRR528 pKa = 11.84SDD530 pKa = 3.54APTAPIRR537 pKa = 11.84VPGLSQRR544 pKa = 11.84EE545 pKa = 3.86NKK547 pKa = 9.9AALRR551 pKa = 11.84AFKK554 pKa = 10.26ILNPRR559 pKa = 11.84TFTDD563 pKa = 2.76AMLRR567 pKa = 11.84AGRR570 pKa = 11.84PTPLTTVQRR579 pKa = 11.84RR580 pKa = 11.84GLLVLKK586 pKa = 10.39NAAPSSGARR595 pKa = 11.84SLWKK599 pKa = 10.46SSLAKK604 pKa = 9.75RR605 pKa = 11.84GHH607 pKa = 5.85YY608 pKa = 10.06ASLMTSYY615 pKa = 10.65LRR617 pKa = 11.84QLMDD621 pKa = 3.75INLPACSKK629 pKa = 11.0APVNGSYY636 pKa = 10.23LARR639 pKa = 11.84MSGTVSQLVTARR651 pKa = 11.84LNLPKK656 pKa = 10.08DD657 pKa = 3.01SRR659 pKa = 11.84MILRR663 pKa = 11.84PSYY666 pKa = 10.35VLSRR670 pKa = 11.84LGKK673 pKa = 9.56RR674 pKa = 11.84LDD676 pKa = 3.88KK677 pKa = 11.04KK678 pKa = 10.06MHH680 pKa = 5.82TPKK683 pKa = 10.3QSSVLRR689 pKa = 11.84LLRR692 pKa = 11.84GLEE695 pKa = 4.01AEE697 pKa = 4.54MVPPSHH703 pKa = 6.63VSVMEE708 pKa = 3.76KK709 pKa = 10.5LLRR712 pKa = 11.84SMVGPDD718 pKa = 3.29DD719 pKa = 3.73GARR722 pKa = 11.84PISGVIGDD730 pKa = 4.34FSQLGITSPTEE741 pKa = 3.92LYY743 pKa = 10.1PQFLHH748 pKa = 6.71WIEE751 pKa = 4.43EE752 pKa = 4.44GAPP755 pKa = 3.36
MM1 pKa = 7.99PLQSMPSQKK10 pKa = 10.66VNFGSLSAVAASLRR24 pKa = 11.84RR25 pKa = 11.84LARR28 pKa = 11.84EE29 pKa = 3.56GRR31 pKa = 11.84TVGSSQYY38 pKa = 10.36ATFARR43 pKa = 11.84LAEE46 pKa = 4.3KK47 pKa = 10.7LRR49 pKa = 11.84GLSPRR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84CLSTASWVAGGLSVSTTDD76 pKa = 3.2RR77 pKa = 11.84SKK79 pKa = 11.35GSSLGHH85 pKa = 6.33RR86 pKa = 11.84LAVVKK91 pKa = 9.18FAIKK95 pKa = 10.01LWNNIISDD103 pKa = 3.89VEE105 pKa = 4.25QFVVDD110 pKa = 3.87YY111 pKa = 11.28KK112 pKa = 10.7RR113 pKa = 11.84VCNSAAYY120 pKa = 9.64AAVRR124 pKa = 11.84GYY126 pKa = 9.62TSIVAAGKK134 pKa = 10.27CRR136 pKa = 11.84DD137 pKa = 3.62YY138 pKa = 11.27LHH140 pKa = 7.24RR141 pKa = 11.84AFGPCGIIPHH151 pKa = 6.69KK152 pKa = 10.52GRR154 pKa = 11.84RR155 pKa = 11.84SLFALASTTRR165 pKa = 11.84CLPPASLTPSRR176 pKa = 11.84VKK178 pKa = 10.49IEE180 pKa = 4.0LEE182 pKa = 3.84ALKK185 pKa = 10.83SRR187 pKa = 11.84LTSVPPPVSGALLSEE202 pKa = 4.36LVSFSHH208 pKa = 7.63DD209 pKa = 3.69LLRR212 pKa = 11.84DD213 pKa = 3.69AGPPVQVEE221 pKa = 4.32VTGNALGPPGKK232 pKa = 9.05TSLDD236 pKa = 3.28RR237 pKa = 11.84AQLYY241 pKa = 10.44GSYY244 pKa = 10.42AFKK247 pKa = 10.77GVIMSKK253 pKa = 8.76TLLMMSKK260 pKa = 10.11SAIPPDD266 pKa = 3.69VPFKK270 pKa = 11.03EE271 pKa = 4.33DD272 pKa = 3.47LEE274 pKa = 4.75EE275 pKa = 4.03NVQRR279 pKa = 11.84VSLLAEE285 pKa = 4.65AGDD288 pKa = 3.83KK289 pKa = 10.57VRR291 pKa = 11.84AITVPSSLAVQLAGRR306 pKa = 11.84SINRR310 pKa = 11.84ALIRR314 pKa = 11.84CLRR317 pKa = 11.84RR318 pKa = 11.84SDD320 pKa = 3.46VFGPGFGKK328 pKa = 10.66RR329 pKa = 11.84PGTLMSGANLGDD341 pKa = 3.56KK342 pKa = 10.68HH343 pKa = 7.46IVSCDD348 pKa = 3.02LTAASDD354 pKa = 3.94YY355 pKa = 11.12LNQDD359 pKa = 2.83VAIAVLADD367 pKa = 3.47YY368 pKa = 10.59LRR370 pKa = 11.84SHH372 pKa = 7.36ASGIWYY378 pKa = 8.48PWASALLGPQRR389 pKa = 11.84IRR391 pKa = 11.84SRR393 pKa = 11.84DD394 pKa = 3.41DD395 pKa = 3.11TYY397 pKa = 8.57TTSRR401 pKa = 11.84GVLMGTPLAWPILSICHH418 pKa = 6.79AFACYY423 pKa = 9.55KK424 pKa = 10.81ALGNTYY430 pKa = 10.43QFLCRR435 pKa = 11.84IVGDD439 pKa = 4.64DD440 pKa = 4.77GLLICNASQYY450 pKa = 9.63RR451 pKa = 11.84AYY453 pKa = 10.61VSVMNRR459 pKa = 11.84LGFVINEE466 pKa = 4.06HH467 pKa = 6.19KK468 pKa = 9.68TFQGKK473 pKa = 8.52QCGILAGRR481 pKa = 11.84LYY483 pKa = 10.81VLKK486 pKa = 9.86TRR488 pKa = 11.84RR489 pKa = 11.84AKK491 pKa = 10.62FQDD494 pKa = 3.58LNSPTKK500 pKa = 8.93TMAWRR505 pKa = 11.84MVDD508 pKa = 3.32CQAPYY513 pKa = 10.74VPEE516 pKa = 4.05LLAARR521 pKa = 11.84GAQRR525 pKa = 11.84FGRR528 pKa = 11.84SDD530 pKa = 3.54APTAPIRR537 pKa = 11.84VPGLSQRR544 pKa = 11.84EE545 pKa = 3.86NKK547 pKa = 9.9AALRR551 pKa = 11.84AFKK554 pKa = 10.26ILNPRR559 pKa = 11.84TFTDD563 pKa = 2.76AMLRR567 pKa = 11.84AGRR570 pKa = 11.84PTPLTTVQRR579 pKa = 11.84RR580 pKa = 11.84GLLVLKK586 pKa = 10.39NAAPSSGARR595 pKa = 11.84SLWKK599 pKa = 10.46SSLAKK604 pKa = 9.75RR605 pKa = 11.84GHH607 pKa = 5.85YY608 pKa = 10.06ASLMTSYY615 pKa = 10.65LRR617 pKa = 11.84QLMDD621 pKa = 3.75INLPACSKK629 pKa = 11.0APVNGSYY636 pKa = 10.23LARR639 pKa = 11.84MSGTVSQLVTARR651 pKa = 11.84LNLPKK656 pKa = 10.08DD657 pKa = 3.01SRR659 pKa = 11.84MILRR663 pKa = 11.84PSYY666 pKa = 10.35VLSRR670 pKa = 11.84LGKK673 pKa = 9.56RR674 pKa = 11.84LDD676 pKa = 3.88KK677 pKa = 11.04KK678 pKa = 10.06MHH680 pKa = 5.82TPKK683 pKa = 10.3QSSVLRR689 pKa = 11.84LLRR692 pKa = 11.84GLEE695 pKa = 4.01AEE697 pKa = 4.54MVPPSHH703 pKa = 6.63VSVMEE708 pKa = 3.76KK709 pKa = 10.5LLRR712 pKa = 11.84SMVGPDD718 pKa = 3.29DD719 pKa = 3.73GARR722 pKa = 11.84PISGVIGDD730 pKa = 4.34FSQLGITSPTEE741 pKa = 3.92LYY743 pKa = 10.1PQFLHH748 pKa = 6.71WIEE751 pKa = 4.43EE752 pKa = 4.44GAPP755 pKa = 3.36
Molecular weight: 82.38 kDa
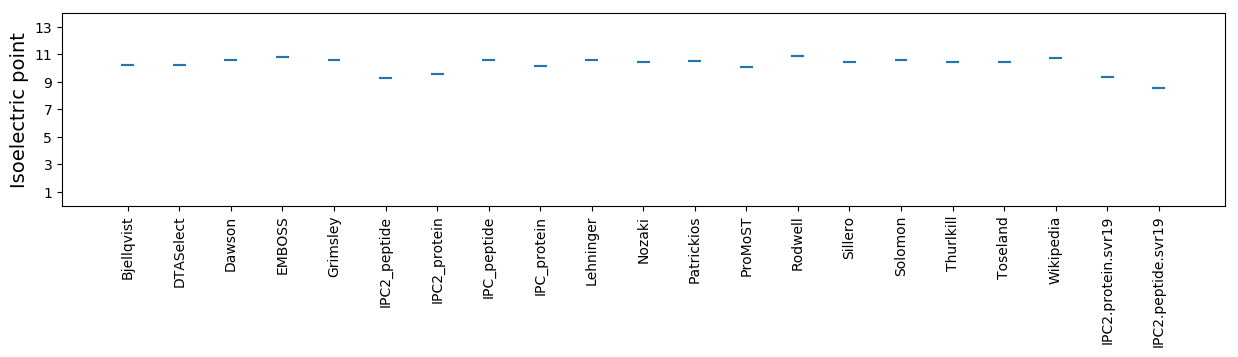
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIK1|A0A1L3KIK1_9VIRU RNA-dependent RNA polymerase OS=Sanxia water strider virus 13 OX=1923397 PE=4 SV=1
MM1 pKa = 7.99PLQSMPSQKK10 pKa = 10.66VNFGSLSAVAASLRR24 pKa = 11.84RR25 pKa = 11.84LARR28 pKa = 11.84EE29 pKa = 3.56GRR31 pKa = 11.84TVGSSQYY38 pKa = 10.36ATFARR43 pKa = 11.84LAEE46 pKa = 4.3KK47 pKa = 10.7LRR49 pKa = 11.84GLSPRR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84CLSTASWVAGGLSVSTTDD76 pKa = 3.2RR77 pKa = 11.84SKK79 pKa = 11.35GSSLGHH85 pKa = 6.33RR86 pKa = 11.84LAVVKK91 pKa = 9.18FAIKK95 pKa = 10.01LWNNIISDD103 pKa = 3.89VEE105 pKa = 4.25QFVVDD110 pKa = 3.87YY111 pKa = 11.28KK112 pKa = 10.7RR113 pKa = 11.84VCNSAAYY120 pKa = 9.64AAVRR124 pKa = 11.84GYY126 pKa = 9.62TSIVAAGKK134 pKa = 10.27CRR136 pKa = 11.84DD137 pKa = 3.62YY138 pKa = 11.27LHH140 pKa = 7.24RR141 pKa = 11.84AFGPCGIIPHH151 pKa = 6.69KK152 pKa = 10.52GRR154 pKa = 11.84RR155 pKa = 11.84SLFALASTTRR165 pKa = 11.84CLPPASLTPSRR176 pKa = 11.84VKK178 pKa = 10.49IEE180 pKa = 4.0LEE182 pKa = 3.84ALKK185 pKa = 10.83SRR187 pKa = 11.84LTSVPPPVSGALLSEE202 pKa = 4.36LVSFSHH208 pKa = 7.63DD209 pKa = 3.69LLRR212 pKa = 11.84DD213 pKa = 3.69AGPPVQVEE221 pKa = 4.32VTGNALGPPGKK232 pKa = 9.05TSLDD236 pKa = 3.28RR237 pKa = 11.84AQLYY241 pKa = 10.44GSYY244 pKa = 10.42AFKK247 pKa = 10.77GVIMSKK253 pKa = 8.76TLLMMSKK260 pKa = 10.11SAIPPDD266 pKa = 3.69VPFKK270 pKa = 11.03EE271 pKa = 4.33DD272 pKa = 3.47LEE274 pKa = 4.75EE275 pKa = 4.03NVQRR279 pKa = 11.84VSLLAEE285 pKa = 4.65AGDD288 pKa = 3.83KK289 pKa = 10.57VRR291 pKa = 11.84AITVPSSLAVQLAGRR306 pKa = 11.84SINRR310 pKa = 11.84ALIRR314 pKa = 11.84CLRR317 pKa = 11.84RR318 pKa = 11.84SDD320 pKa = 3.46VFGPGFGKK328 pKa = 10.66RR329 pKa = 11.84PGTLMSGANLGDD341 pKa = 3.56KK342 pKa = 10.68HH343 pKa = 7.46IVSCDD348 pKa = 3.02LTAASDD354 pKa = 3.94YY355 pKa = 11.12LNQDD359 pKa = 2.83VAIAVLADD367 pKa = 3.47YY368 pKa = 10.59LRR370 pKa = 11.84SHH372 pKa = 7.36ASGIWYY378 pKa = 8.48PWASALLGPQRR389 pKa = 11.84IRR391 pKa = 11.84SRR393 pKa = 11.84DD394 pKa = 3.41DD395 pKa = 3.11TYY397 pKa = 8.57TTSRR401 pKa = 11.84GVLMGTPLAWPILSICHH418 pKa = 6.79AFACYY423 pKa = 9.55KK424 pKa = 10.81ALGNTYY430 pKa = 10.43QFLCRR435 pKa = 11.84IVGDD439 pKa = 4.64DD440 pKa = 4.77GLLICNASQYY450 pKa = 9.63RR451 pKa = 11.84AYY453 pKa = 10.61VSVMNRR459 pKa = 11.84LGFVINEE466 pKa = 4.06HH467 pKa = 6.19KK468 pKa = 9.68TFQGKK473 pKa = 8.52QCGILAGRR481 pKa = 11.84LYY483 pKa = 10.81VLKK486 pKa = 9.86TRR488 pKa = 11.84RR489 pKa = 11.84AKK491 pKa = 10.62FQDD494 pKa = 3.58LNSPTKK500 pKa = 8.93TMAWRR505 pKa = 11.84MVDD508 pKa = 3.32CQAPYY513 pKa = 10.74VPEE516 pKa = 4.05LLAARR521 pKa = 11.84GAQRR525 pKa = 11.84FGRR528 pKa = 11.84SDD530 pKa = 3.54APTAPIRR537 pKa = 11.84VPGLSQRR544 pKa = 11.84EE545 pKa = 3.86NKK547 pKa = 9.9AALRR551 pKa = 11.84AFKK554 pKa = 10.26ILNPRR559 pKa = 11.84TFTDD563 pKa = 2.76AMLRR567 pKa = 11.84AGRR570 pKa = 11.84PTPLTTVQRR579 pKa = 11.84RR580 pKa = 11.84GLLVLKK586 pKa = 10.39NAAPSSGARR595 pKa = 11.84SLWKK599 pKa = 10.46SSLAKK604 pKa = 9.75RR605 pKa = 11.84GHH607 pKa = 5.85YY608 pKa = 10.06ASLMTSYY615 pKa = 10.65LRR617 pKa = 11.84QLMDD621 pKa = 3.75INLPACSKK629 pKa = 11.0APVNGSYY636 pKa = 10.23LARR639 pKa = 11.84MSGTVSQLVTARR651 pKa = 11.84LNLPKK656 pKa = 10.08DD657 pKa = 3.01SRR659 pKa = 11.84MILRR663 pKa = 11.84PSYY666 pKa = 10.35VLSRR670 pKa = 11.84LGKK673 pKa = 9.56RR674 pKa = 11.84LDD676 pKa = 3.88KK677 pKa = 11.04KK678 pKa = 10.06MHH680 pKa = 5.82TPKK683 pKa = 10.3QSSVLRR689 pKa = 11.84LLRR692 pKa = 11.84GLEE695 pKa = 4.01AEE697 pKa = 4.54MVPPSHH703 pKa = 6.63VSVMEE708 pKa = 3.76KK709 pKa = 10.5LLRR712 pKa = 11.84SMVGPDD718 pKa = 3.29DD719 pKa = 3.73GARR722 pKa = 11.84PISGVIGDD730 pKa = 4.34FSQLGITSPTEE741 pKa = 3.92LYY743 pKa = 10.1PQFLHH748 pKa = 6.71WIEE751 pKa = 4.43EE752 pKa = 4.44GAPP755 pKa = 3.36
MM1 pKa = 7.99PLQSMPSQKK10 pKa = 10.66VNFGSLSAVAASLRR24 pKa = 11.84RR25 pKa = 11.84LARR28 pKa = 11.84EE29 pKa = 3.56GRR31 pKa = 11.84TVGSSQYY38 pKa = 10.36ATFARR43 pKa = 11.84LAEE46 pKa = 4.3KK47 pKa = 10.7LRR49 pKa = 11.84GLSPRR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84CLSTASWVAGGLSVSTTDD76 pKa = 3.2RR77 pKa = 11.84SKK79 pKa = 11.35GSSLGHH85 pKa = 6.33RR86 pKa = 11.84LAVVKK91 pKa = 9.18FAIKK95 pKa = 10.01LWNNIISDD103 pKa = 3.89VEE105 pKa = 4.25QFVVDD110 pKa = 3.87YY111 pKa = 11.28KK112 pKa = 10.7RR113 pKa = 11.84VCNSAAYY120 pKa = 9.64AAVRR124 pKa = 11.84GYY126 pKa = 9.62TSIVAAGKK134 pKa = 10.27CRR136 pKa = 11.84DD137 pKa = 3.62YY138 pKa = 11.27LHH140 pKa = 7.24RR141 pKa = 11.84AFGPCGIIPHH151 pKa = 6.69KK152 pKa = 10.52GRR154 pKa = 11.84RR155 pKa = 11.84SLFALASTTRR165 pKa = 11.84CLPPASLTPSRR176 pKa = 11.84VKK178 pKa = 10.49IEE180 pKa = 4.0LEE182 pKa = 3.84ALKK185 pKa = 10.83SRR187 pKa = 11.84LTSVPPPVSGALLSEE202 pKa = 4.36LVSFSHH208 pKa = 7.63DD209 pKa = 3.69LLRR212 pKa = 11.84DD213 pKa = 3.69AGPPVQVEE221 pKa = 4.32VTGNALGPPGKK232 pKa = 9.05TSLDD236 pKa = 3.28RR237 pKa = 11.84AQLYY241 pKa = 10.44GSYY244 pKa = 10.42AFKK247 pKa = 10.77GVIMSKK253 pKa = 8.76TLLMMSKK260 pKa = 10.11SAIPPDD266 pKa = 3.69VPFKK270 pKa = 11.03EE271 pKa = 4.33DD272 pKa = 3.47LEE274 pKa = 4.75EE275 pKa = 4.03NVQRR279 pKa = 11.84VSLLAEE285 pKa = 4.65AGDD288 pKa = 3.83KK289 pKa = 10.57VRR291 pKa = 11.84AITVPSSLAVQLAGRR306 pKa = 11.84SINRR310 pKa = 11.84ALIRR314 pKa = 11.84CLRR317 pKa = 11.84RR318 pKa = 11.84SDD320 pKa = 3.46VFGPGFGKK328 pKa = 10.66RR329 pKa = 11.84PGTLMSGANLGDD341 pKa = 3.56KK342 pKa = 10.68HH343 pKa = 7.46IVSCDD348 pKa = 3.02LTAASDD354 pKa = 3.94YY355 pKa = 11.12LNQDD359 pKa = 2.83VAIAVLADD367 pKa = 3.47YY368 pKa = 10.59LRR370 pKa = 11.84SHH372 pKa = 7.36ASGIWYY378 pKa = 8.48PWASALLGPQRR389 pKa = 11.84IRR391 pKa = 11.84SRR393 pKa = 11.84DD394 pKa = 3.41DD395 pKa = 3.11TYY397 pKa = 8.57TTSRR401 pKa = 11.84GVLMGTPLAWPILSICHH418 pKa = 6.79AFACYY423 pKa = 9.55KK424 pKa = 10.81ALGNTYY430 pKa = 10.43QFLCRR435 pKa = 11.84IVGDD439 pKa = 4.64DD440 pKa = 4.77GLLICNASQYY450 pKa = 9.63RR451 pKa = 11.84AYY453 pKa = 10.61VSVMNRR459 pKa = 11.84LGFVINEE466 pKa = 4.06HH467 pKa = 6.19KK468 pKa = 9.68TFQGKK473 pKa = 8.52QCGILAGRR481 pKa = 11.84LYY483 pKa = 10.81VLKK486 pKa = 9.86TRR488 pKa = 11.84RR489 pKa = 11.84AKK491 pKa = 10.62FQDD494 pKa = 3.58LNSPTKK500 pKa = 8.93TMAWRR505 pKa = 11.84MVDD508 pKa = 3.32CQAPYY513 pKa = 10.74VPEE516 pKa = 4.05LLAARR521 pKa = 11.84GAQRR525 pKa = 11.84FGRR528 pKa = 11.84SDD530 pKa = 3.54APTAPIRR537 pKa = 11.84VPGLSQRR544 pKa = 11.84EE545 pKa = 3.86NKK547 pKa = 9.9AALRR551 pKa = 11.84AFKK554 pKa = 10.26ILNPRR559 pKa = 11.84TFTDD563 pKa = 2.76AMLRR567 pKa = 11.84AGRR570 pKa = 11.84PTPLTTVQRR579 pKa = 11.84RR580 pKa = 11.84GLLVLKK586 pKa = 10.39NAAPSSGARR595 pKa = 11.84SLWKK599 pKa = 10.46SSLAKK604 pKa = 9.75RR605 pKa = 11.84GHH607 pKa = 5.85YY608 pKa = 10.06ASLMTSYY615 pKa = 10.65LRR617 pKa = 11.84QLMDD621 pKa = 3.75INLPACSKK629 pKa = 11.0APVNGSYY636 pKa = 10.23LARR639 pKa = 11.84MSGTVSQLVTARR651 pKa = 11.84LNLPKK656 pKa = 10.08DD657 pKa = 3.01SRR659 pKa = 11.84MILRR663 pKa = 11.84PSYY666 pKa = 10.35VLSRR670 pKa = 11.84LGKK673 pKa = 9.56RR674 pKa = 11.84LDD676 pKa = 3.88KK677 pKa = 11.04KK678 pKa = 10.06MHH680 pKa = 5.82TPKK683 pKa = 10.3QSSVLRR689 pKa = 11.84LLRR692 pKa = 11.84GLEE695 pKa = 4.01AEE697 pKa = 4.54MVPPSHH703 pKa = 6.63VSVMEE708 pKa = 3.76KK709 pKa = 10.5LLRR712 pKa = 11.84SMVGPDD718 pKa = 3.29DD719 pKa = 3.73GARR722 pKa = 11.84PISGVIGDD730 pKa = 4.34FSQLGITSPTEE741 pKa = 3.92LYY743 pKa = 10.1PQFLHH748 pKa = 6.71WIEE751 pKa = 4.43EE752 pKa = 4.44GAPP755 pKa = 3.36
Molecular weight: 82.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
755 |
755 |
755 |
755.0 |
82.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.934 ± 0.0 | 1.854 ± 0.0 |
3.974 ± 0.0 | 2.649 ± 0.0 |
2.781 ± 0.0 | 7.417 ± 0.0 |
1.589 ± 0.0 | 3.974 ± 0.0 |
4.768 ± 0.0 | 11.921 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.517 ± 0.0 | 2.649 ± 0.0 |
6.225 ± 0.0 | 3.311 ± 0.0 |
8.874 ± 0.0 | 9.801 ± 0.0 |
4.901 ± 0.0 | 6.887 ± 0.0 |
1.06 ± 0.0 | 2.914 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
