
Human fecal virus Jorvi3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.82
Get precalculated fractions of proteins
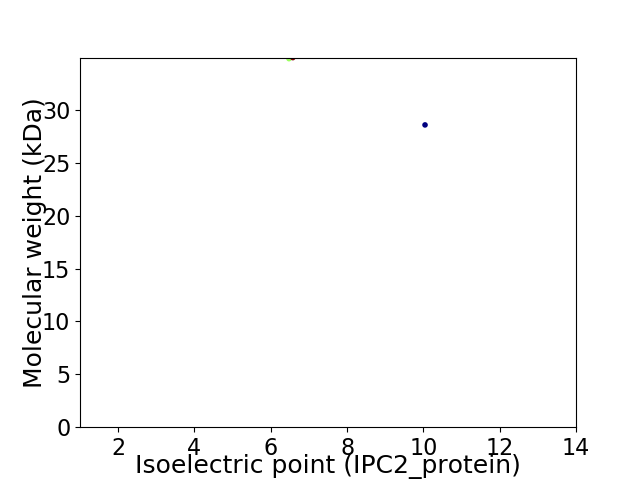
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A220IGD8|A0A220IGD8_9CIRC ATP-dependent helicase Rep OS=Human fecal virus Jorvi3 OX=2017082 PE=3 SV=1
MM1 pKa = 6.71STAYY5 pKa = 10.44CFTLNNPEE13 pKa = 4.02TYY15 pKa = 10.73LPDD18 pKa = 3.85FRR20 pKa = 11.84THH22 pKa = 7.6DD23 pKa = 3.68LSQYY27 pKa = 9.55TNIRR31 pKa = 11.84YY32 pKa = 9.36CIYY35 pKa = 10.12QLEE38 pKa = 4.62EE39 pKa = 3.91GTTPHH44 pKa = 5.98LQGYY48 pKa = 8.16LQLSRR53 pKa = 11.84IQKK56 pKa = 9.72IINIQKK62 pKa = 9.39QEE64 pKa = 3.99FFKK67 pKa = 10.9HH68 pKa = 5.55CSFRR72 pKa = 11.84QANGTPEE79 pKa = 3.88QNIVYY84 pKa = 9.85CSKK87 pKa = 10.76KK88 pKa = 10.59DD89 pKa = 3.21EE90 pKa = 4.54TYY92 pKa = 10.82RR93 pKa = 11.84EE94 pKa = 4.41GPWIYY99 pKa = 9.85GTIKK103 pKa = 10.37QQGKK107 pKa = 10.38RR108 pKa = 11.84NDD110 pKa = 4.1LEE112 pKa = 4.73NMAEE116 pKa = 4.54DD117 pKa = 3.16ILVNKK122 pKa = 8.22KK123 pKa = 8.1TRR125 pKa = 11.84LEE127 pKa = 3.85IAKK130 pKa = 9.08EE131 pKa = 3.89YY132 pKa = 9.07PANFMRR138 pKa = 11.84YY139 pKa = 8.45PRR141 pKa = 11.84GINALFDD148 pKa = 3.76LAIPKK153 pKa = 9.78RR154 pKa = 11.84DD155 pKa = 3.5FKK157 pKa = 11.29TEE159 pKa = 3.62VIYY162 pKa = 10.26IYY164 pKa = 10.86GSTGTGKK171 pKa = 10.08SKK173 pKa = 10.47YY174 pKa = 10.11AHH176 pKa = 6.11QFDD179 pKa = 4.09KK180 pKa = 11.28NAYY183 pKa = 6.65TKK185 pKa = 10.88SFDD188 pKa = 3.85KK189 pKa = 10.4WWDD192 pKa = 3.36QYY194 pKa = 11.77NGQDD198 pKa = 3.47TVIMDD203 pKa = 4.78DD204 pKa = 3.57YY205 pKa = 11.3HH206 pKa = 9.09GEE208 pKa = 3.9LSFNQLLALTDD219 pKa = 3.73RR220 pKa = 11.84YY221 pKa = 10.4EE222 pKa = 4.29CRR224 pKa = 11.84IEE226 pKa = 3.97YY227 pKa = 10.02KK228 pKa = 10.74GSSLQFNSKK237 pKa = 10.07YY238 pKa = 10.82LFIISTRR245 pKa = 11.84LPDD248 pKa = 3.21DD249 pKa = 4.49WYY251 pKa = 10.63PEE253 pKa = 3.85ISPLRR258 pKa = 11.84MEE260 pKa = 4.2EE261 pKa = 4.17FYY263 pKa = 11.23RR264 pKa = 11.84RR265 pKa = 11.84IDD267 pKa = 3.29TVIYY271 pKa = 8.5FTEE274 pKa = 4.21KK275 pKa = 9.35EE276 pKa = 4.19QYY278 pKa = 10.17QEE280 pKa = 4.02FHH282 pKa = 6.86SFSCFKK288 pKa = 10.83KK289 pKa = 10.51III291 pKa = 3.87
MM1 pKa = 6.71STAYY5 pKa = 10.44CFTLNNPEE13 pKa = 4.02TYY15 pKa = 10.73LPDD18 pKa = 3.85FRR20 pKa = 11.84THH22 pKa = 7.6DD23 pKa = 3.68LSQYY27 pKa = 9.55TNIRR31 pKa = 11.84YY32 pKa = 9.36CIYY35 pKa = 10.12QLEE38 pKa = 4.62EE39 pKa = 3.91GTTPHH44 pKa = 5.98LQGYY48 pKa = 8.16LQLSRR53 pKa = 11.84IQKK56 pKa = 9.72IINIQKK62 pKa = 9.39QEE64 pKa = 3.99FFKK67 pKa = 10.9HH68 pKa = 5.55CSFRR72 pKa = 11.84QANGTPEE79 pKa = 3.88QNIVYY84 pKa = 9.85CSKK87 pKa = 10.76KK88 pKa = 10.59DD89 pKa = 3.21EE90 pKa = 4.54TYY92 pKa = 10.82RR93 pKa = 11.84EE94 pKa = 4.41GPWIYY99 pKa = 9.85GTIKK103 pKa = 10.37QQGKK107 pKa = 10.38RR108 pKa = 11.84NDD110 pKa = 4.1LEE112 pKa = 4.73NMAEE116 pKa = 4.54DD117 pKa = 3.16ILVNKK122 pKa = 8.22KK123 pKa = 8.1TRR125 pKa = 11.84LEE127 pKa = 3.85IAKK130 pKa = 9.08EE131 pKa = 3.89YY132 pKa = 9.07PANFMRR138 pKa = 11.84YY139 pKa = 8.45PRR141 pKa = 11.84GINALFDD148 pKa = 3.76LAIPKK153 pKa = 9.78RR154 pKa = 11.84DD155 pKa = 3.5FKK157 pKa = 11.29TEE159 pKa = 3.62VIYY162 pKa = 10.26IYY164 pKa = 10.86GSTGTGKK171 pKa = 10.08SKK173 pKa = 10.47YY174 pKa = 10.11AHH176 pKa = 6.11QFDD179 pKa = 4.09KK180 pKa = 11.28NAYY183 pKa = 6.65TKK185 pKa = 10.88SFDD188 pKa = 3.85KK189 pKa = 10.4WWDD192 pKa = 3.36QYY194 pKa = 11.77NGQDD198 pKa = 3.47TVIMDD203 pKa = 4.78DD204 pKa = 3.57YY205 pKa = 11.3HH206 pKa = 9.09GEE208 pKa = 3.9LSFNQLLALTDD219 pKa = 3.73RR220 pKa = 11.84YY221 pKa = 10.4EE222 pKa = 4.29CRR224 pKa = 11.84IEE226 pKa = 3.97YY227 pKa = 10.02KK228 pKa = 10.74GSSLQFNSKK237 pKa = 10.07YY238 pKa = 10.82LFIISTRR245 pKa = 11.84LPDD248 pKa = 3.21DD249 pKa = 4.49WYY251 pKa = 10.63PEE253 pKa = 3.85ISPLRR258 pKa = 11.84MEE260 pKa = 4.2EE261 pKa = 4.17FYY263 pKa = 11.23RR264 pKa = 11.84RR265 pKa = 11.84IDD267 pKa = 3.29TVIYY271 pKa = 8.5FTEE274 pKa = 4.21KK275 pKa = 9.35EE276 pKa = 4.19QYY278 pKa = 10.17QEE280 pKa = 4.02FHH282 pKa = 6.86SFSCFKK288 pKa = 10.83KK289 pKa = 10.51III291 pKa = 3.87
Molecular weight: 34.84 kDa
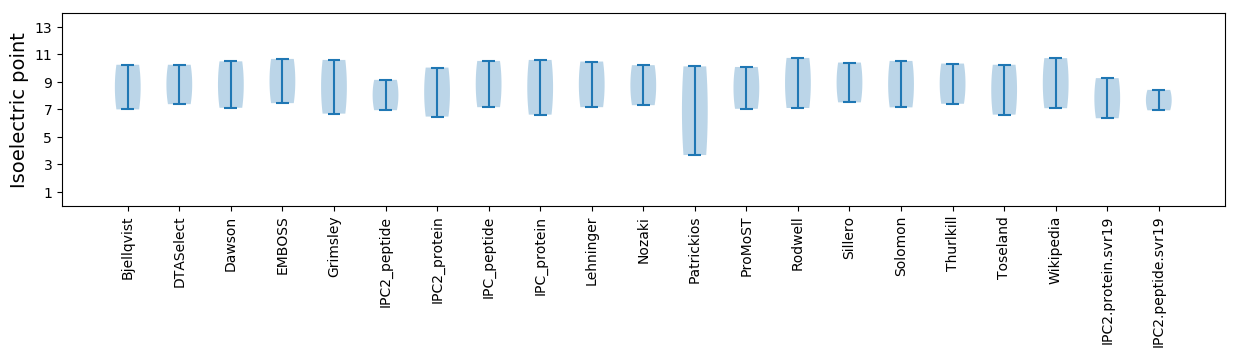
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGD8|A0A220IGD8_9CIRC ATP-dependent helicase Rep OS=Human fecal virus Jorvi3 OX=2017082 PE=3 SV=1
MM1 pKa = 7.79PGFKK5 pKa = 9.67RR6 pKa = 11.84SRR8 pKa = 11.84RR9 pKa = 11.84LYY11 pKa = 9.13SSKK14 pKa = 10.03RR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 8.99GRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84TFKK24 pKa = 10.84KK25 pKa = 9.49VMHH28 pKa = 5.08YY29 pKa = 10.01RR30 pKa = 11.84KK31 pKa = 9.66RR32 pKa = 11.84RR33 pKa = 11.84GAKK36 pKa = 9.63GYY38 pKa = 10.53GRR40 pKa = 11.84ASNLRR45 pKa = 11.84IRR47 pKa = 11.84YY48 pKa = 8.37PLFPDD53 pKa = 3.42RR54 pKa = 11.84VFIKK58 pKa = 10.56LNYY61 pKa = 9.53AEE63 pKa = 5.4RR64 pKa = 11.84INATSTSGGISQYY77 pKa = 10.83LFRR80 pKa = 11.84GNSLFDD86 pKa = 4.57PNFSGTGHH94 pKa = 5.77QPYY97 pKa = 10.34GYY99 pKa = 9.61DD100 pKa = 2.64QWANMYY106 pKa = 10.35SSYY109 pKa = 10.41RR110 pKa = 11.84VHH112 pKa = 6.78GSKK115 pKa = 9.25FTVKK119 pKa = 10.55LLPITDD125 pKa = 4.13TNPEE129 pKa = 3.68IQAAGLMDD137 pKa = 4.29AVLVAFSGINTLSYY151 pKa = 11.25VSIDD155 pKa = 3.5QARR158 pKa = 11.84EE159 pKa = 3.78SPYY162 pKa = 10.78NRR164 pKa = 11.84HH165 pKa = 5.29TLLNRR170 pKa = 11.84NGSRR174 pKa = 11.84QGYY177 pKa = 9.1LKK179 pKa = 10.12MYY181 pKa = 9.28MSTAKK186 pKa = 9.67MYY188 pKa = 10.61GVSKK192 pKa = 11.14VNIQANDD199 pKa = 3.2AYY201 pKa = 10.76AAGTGANPGNQFQWILIVKK220 pKa = 9.81SADD223 pKa = 3.25NLVTITALADD233 pKa = 3.52VKK235 pKa = 9.9ITYY238 pKa = 10.02YY239 pKa = 10.85CEE241 pKa = 3.81LYY243 pKa = 11.01NRR245 pKa = 11.84ISVNQSS251 pKa = 2.3
MM1 pKa = 7.79PGFKK5 pKa = 9.67RR6 pKa = 11.84SRR8 pKa = 11.84RR9 pKa = 11.84LYY11 pKa = 9.13SSKK14 pKa = 10.03RR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 8.99GRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84TFKK24 pKa = 10.84KK25 pKa = 9.49VMHH28 pKa = 5.08YY29 pKa = 10.01RR30 pKa = 11.84KK31 pKa = 9.66RR32 pKa = 11.84RR33 pKa = 11.84GAKK36 pKa = 9.63GYY38 pKa = 10.53GRR40 pKa = 11.84ASNLRR45 pKa = 11.84IRR47 pKa = 11.84YY48 pKa = 8.37PLFPDD53 pKa = 3.42RR54 pKa = 11.84VFIKK58 pKa = 10.56LNYY61 pKa = 9.53AEE63 pKa = 5.4RR64 pKa = 11.84INATSTSGGISQYY77 pKa = 10.83LFRR80 pKa = 11.84GNSLFDD86 pKa = 4.57PNFSGTGHH94 pKa = 5.77QPYY97 pKa = 10.34GYY99 pKa = 9.61DD100 pKa = 2.64QWANMYY106 pKa = 10.35SSYY109 pKa = 10.41RR110 pKa = 11.84VHH112 pKa = 6.78GSKK115 pKa = 9.25FTVKK119 pKa = 10.55LLPITDD125 pKa = 4.13TNPEE129 pKa = 3.68IQAAGLMDD137 pKa = 4.29AVLVAFSGINTLSYY151 pKa = 11.25VSIDD155 pKa = 3.5QARR158 pKa = 11.84EE159 pKa = 3.78SPYY162 pKa = 10.78NRR164 pKa = 11.84HH165 pKa = 5.29TLLNRR170 pKa = 11.84NGSRR174 pKa = 11.84QGYY177 pKa = 9.1LKK179 pKa = 10.12MYY181 pKa = 9.28MSTAKK186 pKa = 9.67MYY188 pKa = 10.61GVSKK192 pKa = 11.14VNIQANDD199 pKa = 3.2AYY201 pKa = 10.76AAGTGANPGNQFQWILIVKK220 pKa = 9.81SADD223 pKa = 3.25NLVTITALADD233 pKa = 3.52VKK235 pKa = 9.9ITYY238 pKa = 10.02YY239 pKa = 10.85CEE241 pKa = 3.81LYY243 pKa = 11.01NRR245 pKa = 11.84ISVNQSS251 pKa = 2.3
Molecular weight: 28.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
542 |
251 |
291 |
271.0 |
31.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.351 ± 1.469 | 1.292 ± 0.591 |
4.797 ± 0.802 | 4.613 ± 1.998 |
5.166 ± 0.782 | 6.089 ± 1.244 |
1.845 ± 0.166 | 7.011 ± 0.949 |
6.642 ± 0.705 | 7.011 ± 0.106 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.214 ± 0.381 | 6.089 ± 0.717 |
3.69 ± 0.069 | 5.166 ± 0.782 |
7.196 ± 1.303 | 7.011 ± 1.161 |
6.273 ± 0.46 | 3.321 ± 1.23 |
1.107 ± 0.205 | 8.118 ± 0.099 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
