
Pelagibacter phage HTVC119P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Votkovvirus; unclassified Votkovvirus
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
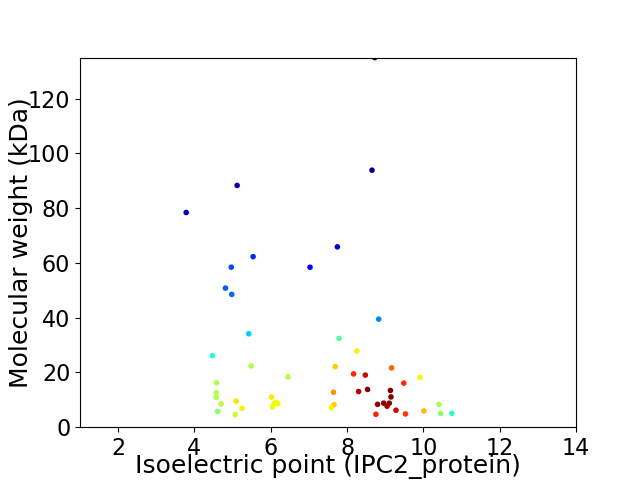
Virtual 2D-PAGE plot for 54 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

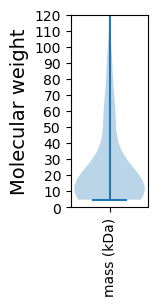
Protein with the lowest isoelectric point:
>tr|A0A4Y1NTN2|A0A4Y1NTN2_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC119P OX=2283020 GN=P119_gp27 PE=4 SV=1
MM1 pKa = 7.57ANSFLEE7 pKa = 4.11YY8 pKa = 9.86TGNGNTTAFSITFDD22 pKa = 3.78YY23 pKa = 11.26LDD25 pKa = 4.44ASHH28 pKa = 7.46IACTVNGVSTSFTLSNGGATANISPAPANAAAIRR62 pKa = 11.84FTRR65 pKa = 11.84TTSQSTRR72 pKa = 11.84LTDD75 pKa = 3.53YY76 pKa = 11.04VAGSVLKK83 pKa = 10.94EE84 pKa = 3.86EE85 pKa = 5.72DD86 pKa = 4.17LDD88 pKa = 4.11TDD90 pKa = 3.91SKK92 pKa = 11.34QAFFMAQEE100 pKa = 4.71GIEE103 pKa = 4.36TISSKK108 pKa = 10.07MGQSTTNFQFDD119 pKa = 3.99ALNKK123 pKa = 10.07RR124 pKa = 11.84IINVADD130 pKa = 3.7PVDD133 pKa = 3.57NTDD136 pKa = 3.33AVNKK140 pKa = 9.92QFISTNLPNITTVAGISSDD159 pKa = 3.43VTTVAGISSDD169 pKa = 3.42VTAVANNEE177 pKa = 3.7ADD179 pKa = 3.43IDD181 pKa = 4.17LVALNMPSVTTVATNINDD199 pKa = 4.02VITVANDD206 pKa = 3.15LNEE209 pKa = 4.92AISEE213 pKa = 4.21IEE215 pKa = 3.93TAANDD220 pKa = 3.8LNEE223 pKa = 4.38ATSEE227 pKa = 3.83IDD229 pKa = 3.41TVSNNIANVNIVGTNIGNVNTIATANTDD257 pKa = 3.16IQTLADD263 pKa = 4.24LEE265 pKa = 5.04DD266 pKa = 4.01GTVTTNGLSTLAGLNTEE283 pKa = 3.89IQGVYY288 pKa = 10.47NIRR291 pKa = 11.84NNVTNVDD298 pKa = 3.67TNATNVNLVAGQISPTNNVSAVSAVATEE326 pKa = 3.92IGTLGALGTEE336 pKa = 3.91ITNLNNIRR344 pKa = 11.84TDD346 pKa = 2.93ITGVNTISADD356 pKa = 3.41VTGVNNIAPAVTAVNNNSANINAVNANSVNINAVKK391 pKa = 10.75NNEE394 pKa = 3.94ANINAVNANSANINTVAGLNTEE416 pKa = 4.05ITALGASGTVASINTVANNLASVNSFANTYY446 pKa = 10.15LGASATAPTQDD457 pKa = 3.92PDD459 pKa = 4.51GSALDD464 pKa = 4.82LGDD467 pKa = 5.26LYY469 pKa = 11.34FDD471 pKa = 3.95TASDD475 pKa = 3.54TMKK478 pKa = 10.42VYY480 pKa = 10.97SSGGWINAGSAVNGTANRR498 pKa = 11.84FKK500 pKa = 10.31YY501 pKa = 8.64TATASQTTFSGADD514 pKa = 3.29DD515 pKa = 3.9SGNTLAYY522 pKa = 10.3DD523 pKa = 3.29SGYY526 pKa = 11.19ADD528 pKa = 3.57IYY530 pKa = 10.88LNGVKK535 pKa = 9.97LVNGSDD541 pKa = 3.7FTATSGTSIVLASGASANDD560 pKa = 3.21ILEE563 pKa = 4.43VIAYY567 pKa = 7.63GTFTLSNFSITNANDD582 pKa = 3.06VPALGSAGQVLQVNSGATALEE603 pKa = 4.51FGTVDD608 pKa = 4.95LANLNADD615 pKa = 3.68NLTSGTVPSARR626 pKa = 11.84VSGAYY631 pKa = 8.91TGITQTGTLTKK642 pKa = 10.42SGATVTTFNRR652 pKa = 11.84TTSDD656 pKa = 3.34GNILEE661 pKa = 4.32LQKK664 pKa = 10.51DD665 pKa = 3.79TSLSGSVGSVGGNGAFYY682 pKa = 10.4ISGANNVGLQFNNVNDD698 pKa = 4.43EE699 pKa = 4.0IRR701 pKa = 11.84PCNSDD706 pKa = 2.65GSARR710 pKa = 11.84DD711 pKa = 3.49NAIDD715 pKa = 4.2LGNATRR721 pKa = 11.84RR722 pKa = 11.84FKK724 pKa = 10.88DD725 pKa = 3.36LYY727 pKa = 10.77LGGGLYY733 pKa = 10.72VGGTGTANKK742 pKa = 10.15LDD744 pKa = 4.51DD745 pKa = 4.22YY746 pKa = 11.86EE747 pKa = 4.58EE748 pKa = 4.74GTWTPNVNSTSGFTNTNVV766 pKa = 2.86
MM1 pKa = 7.57ANSFLEE7 pKa = 4.11YY8 pKa = 9.86TGNGNTTAFSITFDD22 pKa = 3.78YY23 pKa = 11.26LDD25 pKa = 4.44ASHH28 pKa = 7.46IACTVNGVSTSFTLSNGGATANISPAPANAAAIRR62 pKa = 11.84FTRR65 pKa = 11.84TTSQSTRR72 pKa = 11.84LTDD75 pKa = 3.53YY76 pKa = 11.04VAGSVLKK83 pKa = 10.94EE84 pKa = 3.86EE85 pKa = 5.72DD86 pKa = 4.17LDD88 pKa = 4.11TDD90 pKa = 3.91SKK92 pKa = 11.34QAFFMAQEE100 pKa = 4.71GIEE103 pKa = 4.36TISSKK108 pKa = 10.07MGQSTTNFQFDD119 pKa = 3.99ALNKK123 pKa = 10.07RR124 pKa = 11.84IINVADD130 pKa = 3.7PVDD133 pKa = 3.57NTDD136 pKa = 3.33AVNKK140 pKa = 9.92QFISTNLPNITTVAGISSDD159 pKa = 3.43VTTVAGISSDD169 pKa = 3.42VTAVANNEE177 pKa = 3.7ADD179 pKa = 3.43IDD181 pKa = 4.17LVALNMPSVTTVATNINDD199 pKa = 4.02VITVANDD206 pKa = 3.15LNEE209 pKa = 4.92AISEE213 pKa = 4.21IEE215 pKa = 3.93TAANDD220 pKa = 3.8LNEE223 pKa = 4.38ATSEE227 pKa = 3.83IDD229 pKa = 3.41TVSNNIANVNIVGTNIGNVNTIATANTDD257 pKa = 3.16IQTLADD263 pKa = 4.24LEE265 pKa = 5.04DD266 pKa = 4.01GTVTTNGLSTLAGLNTEE283 pKa = 3.89IQGVYY288 pKa = 10.47NIRR291 pKa = 11.84NNVTNVDD298 pKa = 3.67TNATNVNLVAGQISPTNNVSAVSAVATEE326 pKa = 3.92IGTLGALGTEE336 pKa = 3.91ITNLNNIRR344 pKa = 11.84TDD346 pKa = 2.93ITGVNTISADD356 pKa = 3.41VTGVNNIAPAVTAVNNNSANINAVNANSVNINAVKK391 pKa = 10.75NNEE394 pKa = 3.94ANINAVNANSANINTVAGLNTEE416 pKa = 4.05ITALGASGTVASINTVANNLASVNSFANTYY446 pKa = 10.15LGASATAPTQDD457 pKa = 3.92PDD459 pKa = 4.51GSALDD464 pKa = 4.82LGDD467 pKa = 5.26LYY469 pKa = 11.34FDD471 pKa = 3.95TASDD475 pKa = 3.54TMKK478 pKa = 10.42VYY480 pKa = 10.97SSGGWINAGSAVNGTANRR498 pKa = 11.84FKK500 pKa = 10.31YY501 pKa = 8.64TATASQTTFSGADD514 pKa = 3.29DD515 pKa = 3.9SGNTLAYY522 pKa = 10.3DD523 pKa = 3.29SGYY526 pKa = 11.19ADD528 pKa = 3.57IYY530 pKa = 10.88LNGVKK535 pKa = 9.97LVNGSDD541 pKa = 3.7FTATSGTSIVLASGASANDD560 pKa = 3.21ILEE563 pKa = 4.43VIAYY567 pKa = 7.63GTFTLSNFSITNANDD582 pKa = 3.06VPALGSAGQVLQVNSGATALEE603 pKa = 4.51FGTVDD608 pKa = 4.95LANLNADD615 pKa = 3.68NLTSGTVPSARR626 pKa = 11.84VSGAYY631 pKa = 8.91TGITQTGTLTKK642 pKa = 10.42SGATVTTFNRR652 pKa = 11.84TTSDD656 pKa = 3.34GNILEE661 pKa = 4.32LQKK664 pKa = 10.51DD665 pKa = 3.79TSLSGSVGSVGGNGAFYY682 pKa = 10.4ISGANNVGLQFNNVNDD698 pKa = 4.43EE699 pKa = 4.0IRR701 pKa = 11.84PCNSDD706 pKa = 2.65GSARR710 pKa = 11.84DD711 pKa = 3.49NAIDD715 pKa = 4.2LGNATRR721 pKa = 11.84RR722 pKa = 11.84FKK724 pKa = 10.88DD725 pKa = 3.36LYY727 pKa = 10.77LGGGLYY733 pKa = 10.72VGGTGTANKK742 pKa = 10.15LDD744 pKa = 4.51DD745 pKa = 4.22YY746 pKa = 11.86EE747 pKa = 4.58EE748 pKa = 4.74GTWTPNVNSTSGFTNTNVV766 pKa = 2.86
Molecular weight: 78.39 kDa
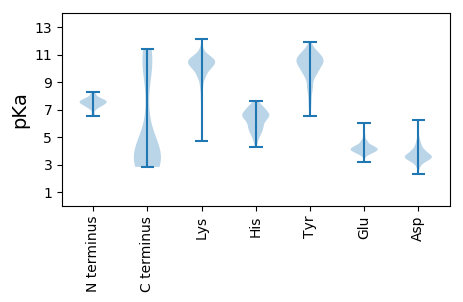
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NT39|A0A4Y1NT39_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC119P OX=2283020 GN=P119_gp24 PE=4 SV=1
MM1 pKa = 7.35ATPGKK6 pKa = 10.38QYY8 pKa = 10.65ISRR11 pKa = 11.84NLSGRR16 pKa = 11.84VLTRR20 pKa = 11.84GYY22 pKa = 9.01TEE24 pKa = 4.73NGWTYY29 pKa = 11.56LDD31 pKa = 3.27LRR33 pKa = 11.84TRR35 pKa = 11.84KK36 pKa = 9.96SRR38 pKa = 11.84VSEE41 pKa = 3.85RR42 pKa = 11.84WVKK45 pKa = 10.5SSYY48 pKa = 10.54FKK50 pKa = 10.87IMCITQAWQASQHH63 pKa = 4.84RR64 pKa = 11.84AKK66 pKa = 10.5RR67 pKa = 11.84KK68 pKa = 9.12KK69 pKa = 9.98LKK71 pKa = 9.17HH72 pKa = 6.05TITLLQLIKK81 pKa = 10.53LYY83 pKa = 9.98PKK85 pKa = 10.91DD86 pKa = 3.48HH87 pKa = 6.51MCPVFKK93 pKa = 10.49TPLVFGGGLNKK104 pKa = 10.11FSPSIDD110 pKa = 3.37RR111 pKa = 11.84INNLKK116 pKa = 10.69GYY118 pKa = 10.51VKK120 pKa = 10.92GNVQWISSRR129 pKa = 11.84ANTLKK134 pKa = 10.42RR135 pKa = 11.84DD136 pKa = 3.58ATAKK140 pKa = 10.14EE141 pKa = 4.81LYY143 pKa = 9.52TLADD147 pKa = 4.06YY148 pKa = 10.91ISTINKK154 pKa = 8.18TKK156 pKa = 10.32II157 pKa = 3.11
MM1 pKa = 7.35ATPGKK6 pKa = 10.38QYY8 pKa = 10.65ISRR11 pKa = 11.84NLSGRR16 pKa = 11.84VLTRR20 pKa = 11.84GYY22 pKa = 9.01TEE24 pKa = 4.73NGWTYY29 pKa = 11.56LDD31 pKa = 3.27LRR33 pKa = 11.84TRR35 pKa = 11.84KK36 pKa = 9.96SRR38 pKa = 11.84VSEE41 pKa = 3.85RR42 pKa = 11.84WVKK45 pKa = 10.5SSYY48 pKa = 10.54FKK50 pKa = 10.87IMCITQAWQASQHH63 pKa = 4.84RR64 pKa = 11.84AKK66 pKa = 10.5RR67 pKa = 11.84KK68 pKa = 9.12KK69 pKa = 9.98LKK71 pKa = 9.17HH72 pKa = 6.05TITLLQLIKK81 pKa = 10.53LYY83 pKa = 9.98PKK85 pKa = 10.91DD86 pKa = 3.48HH87 pKa = 6.51MCPVFKK93 pKa = 10.49TPLVFGGGLNKK104 pKa = 10.11FSPSIDD110 pKa = 3.37RR111 pKa = 11.84INNLKK116 pKa = 10.69GYY118 pKa = 10.51VKK120 pKa = 10.92GNVQWISSRR129 pKa = 11.84ANTLKK134 pKa = 10.42RR135 pKa = 11.84DD136 pKa = 3.58ATAKK140 pKa = 10.14EE141 pKa = 4.81LYY143 pKa = 9.52TLADD147 pKa = 4.06YY148 pKa = 10.91ISTINKK154 pKa = 8.18TKK156 pKa = 10.32II157 pKa = 3.11
Molecular weight: 18.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11891 |
39 |
1210 |
220.2 |
24.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.636 ± 0.433 | 0.95 ± 0.131 |
5.937 ± 0.238 | 6.08 ± 0.404 |
3.734 ± 0.188 | 6.459 ± 0.43 |
1.539 ± 0.186 | 6.921 ± 0.171 |
8.712 ± 0.723 | 8.342 ± 0.341 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.195 ± 0.19 | 6.383 ± 0.485 |
3.271 ± 0.227 | 3.902 ± 0.276 |
4.045 ± 0.28 | 6.787 ± 0.463 |
6.677 ± 0.615 | 5.777 ± 0.245 |
1.118 ± 0.142 | 3.532 ± 0.217 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
