
Fibroporia radiculosa
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporales incertae sedis; Fibroporia
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9251 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
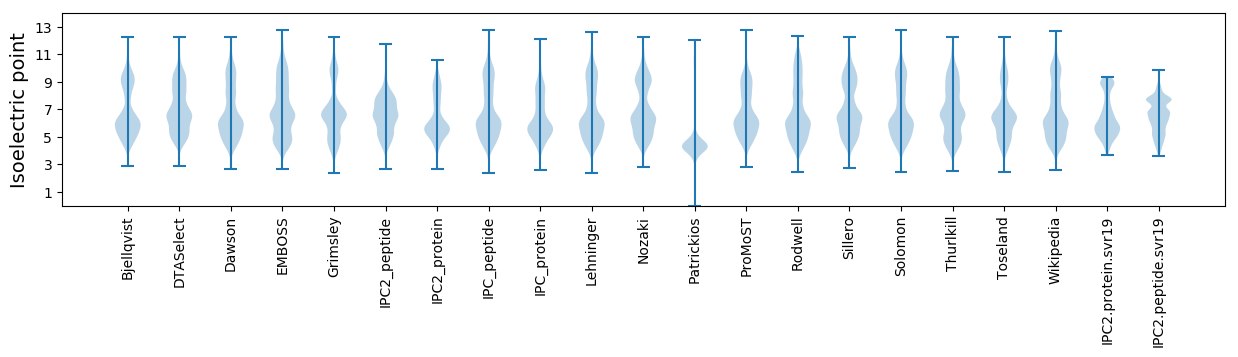
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|J4GN21|J4GN21_9APHY Uncharacterized protein OS=Fibroporia radiculosa OX=599839 GN=FIBRA_02709 PE=4 SV=1
MM1 pKa = 7.2GAYY4 pKa = 9.41YY5 pKa = 10.61DD6 pKa = 3.77EE7 pKa = 5.18VEE9 pKa = 4.36IEE11 pKa = 4.08DD12 pKa = 4.05MAWDD16 pKa = 3.59EE17 pKa = 4.61DD18 pKa = 3.38KK19 pKa = 10.93RR20 pKa = 11.84IYY22 pKa = 10.32HH23 pKa = 5.62YY24 pKa = 9.88PCPCGDD30 pKa = 3.27RR31 pKa = 11.84FEE33 pKa = 4.69ISRR36 pKa = 11.84KK37 pKa = 8.98QLANYY42 pKa = 9.44EE43 pKa = 4.73DD44 pKa = 4.57IATCPSCSLVIRR56 pKa = 11.84VIYY59 pKa = 10.38DD60 pKa = 3.27PLDD63 pKa = 4.13YY64 pKa = 10.93EE65 pKa = 5.51DD66 pKa = 5.26EE67 pKa = 4.91PPDD70 pKa = 4.02EE71 pKa = 5.23DD72 pKa = 5.71DD73 pKa = 4.08EE74 pKa = 5.15GAVSDD79 pKa = 4.5EE80 pKa = 4.72EE81 pKa = 4.72SDD83 pKa = 4.56EE84 pKa = 4.78DD85 pKa = 5.37DD86 pKa = 4.12SDD88 pKa = 4.32SGDD91 pKa = 3.5EE92 pKa = 4.09RR93 pKa = 11.84FEE95 pKa = 4.47DD96 pKa = 4.15ALDD99 pKa = 3.98RR100 pKa = 11.84LHH102 pKa = 7.12LADD105 pKa = 4.73EE106 pKa = 4.8RR107 pKa = 11.84PLVIAVGAA115 pKa = 3.99
MM1 pKa = 7.2GAYY4 pKa = 9.41YY5 pKa = 10.61DD6 pKa = 3.77EE7 pKa = 5.18VEE9 pKa = 4.36IEE11 pKa = 4.08DD12 pKa = 4.05MAWDD16 pKa = 3.59EE17 pKa = 4.61DD18 pKa = 3.38KK19 pKa = 10.93RR20 pKa = 11.84IYY22 pKa = 10.32HH23 pKa = 5.62YY24 pKa = 9.88PCPCGDD30 pKa = 3.27RR31 pKa = 11.84FEE33 pKa = 4.69ISRR36 pKa = 11.84KK37 pKa = 8.98QLANYY42 pKa = 9.44EE43 pKa = 4.73DD44 pKa = 4.57IATCPSCSLVIRR56 pKa = 11.84VIYY59 pKa = 10.38DD60 pKa = 3.27PLDD63 pKa = 4.13YY64 pKa = 10.93EE65 pKa = 5.51DD66 pKa = 5.26EE67 pKa = 4.91PPDD70 pKa = 4.02EE71 pKa = 5.23DD72 pKa = 5.71DD73 pKa = 4.08EE74 pKa = 5.15GAVSDD79 pKa = 4.5EE80 pKa = 4.72EE81 pKa = 4.72SDD83 pKa = 4.56EE84 pKa = 4.78DD85 pKa = 5.37DD86 pKa = 4.12SDD88 pKa = 4.32SGDD91 pKa = 3.5EE92 pKa = 4.09RR93 pKa = 11.84FEE95 pKa = 4.47DD96 pKa = 4.15ALDD99 pKa = 3.98RR100 pKa = 11.84LHH102 pKa = 7.12LADD105 pKa = 4.73EE106 pKa = 4.8RR107 pKa = 11.84PLVIAVGAA115 pKa = 3.99
Molecular weight: 13.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J4HWQ2|J4HWQ2_9APHY Uncharacterized protein OS=Fibroporia radiculosa OX=599839 GN=FIBRA_04761 PE=4 SV=1
MM1 pKa = 7.42SVPRR5 pKa = 11.84LPAIGRR11 pKa = 11.84CSNPCRR17 pKa = 11.84SPWRR21 pKa = 11.84TRR23 pKa = 11.84IQPSLSSPALAALLRR38 pKa = 11.84TRR40 pKa = 11.84TRR42 pKa = 11.84TLTSLPGSSTSSPSSPPPKK61 pKa = 10.49LPMTKK66 pKa = 9.74PLSSPILPQSMPPDD80 pKa = 4.4PDD82 pKa = 4.24FEE84 pKa = 4.71PSPPNSALLLFCLSTGLVDD103 pKa = 4.07SLTFAVSGVWCAFVTGTTVQLGMDD127 pKa = 4.54LPALLAHH134 pKa = 6.6LPALFAHH141 pKa = 7.15LPTSTTEE148 pKa = 3.82LSIATSQFLSSGSSEE163 pKa = 3.92RR164 pKa = 11.84LSALGGFLLGGLSAARR180 pKa = 11.84FPEE183 pKa = 4.95AISPARR189 pKa = 11.84STAFQSALLVMASVLVLTTTPAGAEE214 pKa = 4.01YY215 pKa = 10.57LSYY218 pKa = 11.4SHH220 pKa = 6.89LTLALISSSMAIQAVLVQRR239 pKa = 11.84FATPYY244 pKa = 7.71ATSVAWTSVWLSAVAPGSPTKK265 pKa = 9.72RR266 pKa = 11.84WKK268 pKa = 10.74RR269 pKa = 11.84FVGLGAVMSGAALGASLLVFGVDD292 pKa = 3.7DD293 pKa = 3.91EE294 pKa = 4.61EE295 pKa = 4.6KK296 pKa = 10.54ARR298 pKa = 11.84EE299 pKa = 4.07EE300 pKa = 4.0MVHH303 pKa = 5.46RR304 pKa = 11.84VGWGLAVAALVKK316 pKa = 10.69AIVAGMFWRR325 pKa = 11.84RR326 pKa = 11.84GRR328 pKa = 11.84RR329 pKa = 11.84RR330 pKa = 11.84GIQLL334 pKa = 3.32
MM1 pKa = 7.42SVPRR5 pKa = 11.84LPAIGRR11 pKa = 11.84CSNPCRR17 pKa = 11.84SPWRR21 pKa = 11.84TRR23 pKa = 11.84IQPSLSSPALAALLRR38 pKa = 11.84TRR40 pKa = 11.84TRR42 pKa = 11.84TLTSLPGSSTSSPSSPPPKK61 pKa = 10.49LPMTKK66 pKa = 9.74PLSSPILPQSMPPDD80 pKa = 4.4PDD82 pKa = 4.24FEE84 pKa = 4.71PSPPNSALLLFCLSTGLVDD103 pKa = 4.07SLTFAVSGVWCAFVTGTTVQLGMDD127 pKa = 4.54LPALLAHH134 pKa = 6.6LPALFAHH141 pKa = 7.15LPTSTTEE148 pKa = 3.82LSIATSQFLSSGSSEE163 pKa = 3.92RR164 pKa = 11.84LSALGGFLLGGLSAARR180 pKa = 11.84FPEE183 pKa = 4.95AISPARR189 pKa = 11.84STAFQSALLVMASVLVLTTTPAGAEE214 pKa = 4.01YY215 pKa = 10.57LSYY218 pKa = 11.4SHH220 pKa = 6.89LTLALISSSMAIQAVLVQRR239 pKa = 11.84FATPYY244 pKa = 7.71ATSVAWTSVWLSAVAPGSPTKK265 pKa = 9.72RR266 pKa = 11.84WKK268 pKa = 10.74RR269 pKa = 11.84FVGLGAVMSGAALGASLLVFGVDD292 pKa = 3.7DD293 pKa = 3.91EE294 pKa = 4.61EE295 pKa = 4.6KK296 pKa = 10.54ARR298 pKa = 11.84EE299 pKa = 4.07EE300 pKa = 4.0MVHH303 pKa = 5.46RR304 pKa = 11.84VGWGLAVAALVKK316 pKa = 10.69AIVAGMFWRR325 pKa = 11.84RR326 pKa = 11.84GRR328 pKa = 11.84RR329 pKa = 11.84RR330 pKa = 11.84GIQLL334 pKa = 3.32
Molecular weight: 35.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4412171 |
8 |
5034 |
476.9 |
52.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.808 ± 0.019 | 1.143 ± 0.009 |
5.693 ± 0.019 | 5.876 ± 0.024 |
3.635 ± 0.015 | 6.46 ± 0.023 |
2.523 ± 0.011 | 4.801 ± 0.018 |
4.169 ± 0.022 | 9.331 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.147 ± 0.008 | 3.223 ± 0.012 |
6.548 ± 0.032 | 3.719 ± 0.015 |
6.392 ± 0.024 | 8.937 ± 0.034 |
5.954 ± 0.018 | 6.58 ± 0.018 |
1.396 ± 0.009 | 2.665 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
