
Microviridae Fen685_11
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
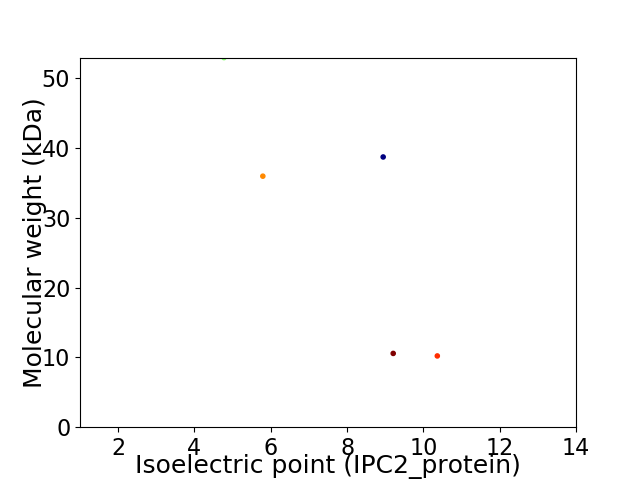
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
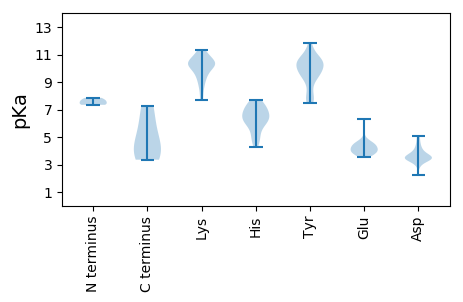
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UMG1|A0A0G2UMG1_9VIRU Major capsid protein VP1 OS=Microviridae Fen685_11 OX=1655657 PE=3 SV=1
MM1 pKa = 7.36QPMLAPVLHH10 pKa = 6.67RR11 pKa = 11.84LSIRR15 pKa = 11.84SEE17 pKa = 3.56WFFIPNRR24 pKa = 11.84ILWPNWEE31 pKa = 4.52SFISPPVEE39 pKa = 4.43GVDD42 pKa = 4.08GPAAPFLSNFTVVSNSLGDD61 pKa = 3.89YY62 pKa = 10.69LGLPLGSWNNGDD74 pKa = 4.46GGDD77 pKa = 3.73PSQIVNALPFSAYY90 pKa = 9.24QRR92 pKa = 11.84VWWDD96 pKa = 2.86WYY98 pKa = 10.54RR99 pKa = 11.84DD100 pKa = 3.34EE101 pKa = 6.28DD102 pKa = 4.08FQPLQWQYY110 pKa = 11.87AGQLVDD116 pKa = 4.39GDD118 pKa = 3.72NSEE121 pKa = 4.16QEE123 pKa = 4.02EE124 pKa = 4.61LFPIRR129 pKa = 11.84IRR131 pKa = 11.84NNDD134 pKa = 2.9RR135 pKa = 11.84DD136 pKa = 4.19YY137 pKa = 10.75FTSAKK142 pKa = 9.64PWPQKK147 pKa = 10.39GDD149 pKa = 3.36AVVIPAFVGDD159 pKa = 4.45LPVSANLEE167 pKa = 4.55TQWHH171 pKa = 5.33VNRR174 pKa = 11.84TDD176 pKa = 3.24HH177 pKa = 6.66TGLSATALVSTGTSDD192 pKa = 3.16TDD194 pKa = 4.71GIGLDD199 pKa = 3.69GGGTGVALDD208 pKa = 3.81PRR210 pKa = 11.84GALSISSDD218 pKa = 4.22DD219 pKa = 3.85MLAQAGTIEE228 pKa = 4.23QLRR231 pKa = 11.84MANALQRR238 pKa = 11.84FLEE241 pKa = 4.42ADD243 pKa = 3.0ARR245 pKa = 11.84GGTRR249 pKa = 11.84YY250 pKa = 9.86VEE252 pKa = 3.94LLWRR256 pKa = 11.84HH257 pKa = 5.84FKK259 pKa = 10.77AYY261 pKa = 10.6LPDD264 pKa = 5.07KK265 pKa = 10.05IDD267 pKa = 3.41RR268 pKa = 11.84SEE270 pKa = 4.34YY271 pKa = 10.15IGNTVQPISISEE283 pKa = 4.1VLNTTGTTDD292 pKa = 3.23APQGTMTGHH301 pKa = 7.14GISAARR307 pKa = 11.84HH308 pKa = 4.88ATDD311 pKa = 4.09LSYY314 pKa = 11.34SAVEE318 pKa = 4.86HH319 pKa = 6.11GWLMCICSVIPATGYY334 pKa = 9.33YY335 pKa = 9.91QGIPKK340 pKa = 8.63MWSRR344 pKa = 11.84FDD346 pKa = 3.53RR347 pKa = 11.84FDD349 pKa = 3.63YY350 pKa = 10.58AWPEE354 pKa = 3.85FAHH357 pKa = 7.12LGEE360 pKa = 4.23QPILNQEE367 pKa = 4.47LYY369 pKa = 10.77YY370 pKa = 10.44DD371 pKa = 4.06TASVDD376 pKa = 3.54PDD378 pKa = 3.63TGNTATWGYY387 pKa = 9.76LPRR390 pKa = 11.84YY391 pKa = 7.63TEE393 pKa = 4.53YY394 pKa = 10.54KK395 pKa = 9.43SQMNRR400 pKa = 11.84VAGDD404 pKa = 3.33FRR406 pKa = 11.84TSLDD410 pKa = 3.3YY411 pKa = 10.5WHH413 pKa = 7.71LARR416 pKa = 11.84KK417 pKa = 8.18FASLPALAQEE427 pKa = 4.58FLEE430 pKa = 4.53ISQGADD436 pKa = 2.82MDD438 pKa = 4.97RR439 pKa = 11.84IFAVIDD445 pKa = 3.32PTVQHH450 pKa = 6.35VLVHH454 pKa = 6.47WFHH457 pKa = 7.14AIKK460 pKa = 10.31VHH462 pKa = 6.35RR463 pKa = 11.84ALPRR467 pKa = 11.84IVTPDD472 pKa = 3.13LL473 pKa = 3.94
MM1 pKa = 7.36QPMLAPVLHH10 pKa = 6.67RR11 pKa = 11.84LSIRR15 pKa = 11.84SEE17 pKa = 3.56WFFIPNRR24 pKa = 11.84ILWPNWEE31 pKa = 4.52SFISPPVEE39 pKa = 4.43GVDD42 pKa = 4.08GPAAPFLSNFTVVSNSLGDD61 pKa = 3.89YY62 pKa = 10.69LGLPLGSWNNGDD74 pKa = 4.46GGDD77 pKa = 3.73PSQIVNALPFSAYY90 pKa = 9.24QRR92 pKa = 11.84VWWDD96 pKa = 2.86WYY98 pKa = 10.54RR99 pKa = 11.84DD100 pKa = 3.34EE101 pKa = 6.28DD102 pKa = 4.08FQPLQWQYY110 pKa = 11.87AGQLVDD116 pKa = 4.39GDD118 pKa = 3.72NSEE121 pKa = 4.16QEE123 pKa = 4.02EE124 pKa = 4.61LFPIRR129 pKa = 11.84IRR131 pKa = 11.84NNDD134 pKa = 2.9RR135 pKa = 11.84DD136 pKa = 4.19YY137 pKa = 10.75FTSAKK142 pKa = 9.64PWPQKK147 pKa = 10.39GDD149 pKa = 3.36AVVIPAFVGDD159 pKa = 4.45LPVSANLEE167 pKa = 4.55TQWHH171 pKa = 5.33VNRR174 pKa = 11.84TDD176 pKa = 3.24HH177 pKa = 6.66TGLSATALVSTGTSDD192 pKa = 3.16TDD194 pKa = 4.71GIGLDD199 pKa = 3.69GGGTGVALDD208 pKa = 3.81PRR210 pKa = 11.84GALSISSDD218 pKa = 4.22DD219 pKa = 3.85MLAQAGTIEE228 pKa = 4.23QLRR231 pKa = 11.84MANALQRR238 pKa = 11.84FLEE241 pKa = 4.42ADD243 pKa = 3.0ARR245 pKa = 11.84GGTRR249 pKa = 11.84YY250 pKa = 9.86VEE252 pKa = 3.94LLWRR256 pKa = 11.84HH257 pKa = 5.84FKK259 pKa = 10.77AYY261 pKa = 10.6LPDD264 pKa = 5.07KK265 pKa = 10.05IDD267 pKa = 3.41RR268 pKa = 11.84SEE270 pKa = 4.34YY271 pKa = 10.15IGNTVQPISISEE283 pKa = 4.1VLNTTGTTDD292 pKa = 3.23APQGTMTGHH301 pKa = 7.14GISAARR307 pKa = 11.84HH308 pKa = 4.88ATDD311 pKa = 4.09LSYY314 pKa = 11.34SAVEE318 pKa = 4.86HH319 pKa = 6.11GWLMCICSVIPATGYY334 pKa = 9.33YY335 pKa = 9.91QGIPKK340 pKa = 8.63MWSRR344 pKa = 11.84FDD346 pKa = 3.53RR347 pKa = 11.84FDD349 pKa = 3.63YY350 pKa = 10.58AWPEE354 pKa = 3.85FAHH357 pKa = 7.12LGEE360 pKa = 4.23QPILNQEE367 pKa = 4.47LYY369 pKa = 10.77YY370 pKa = 10.44DD371 pKa = 4.06TASVDD376 pKa = 3.54PDD378 pKa = 3.63TGNTATWGYY387 pKa = 9.76LPRR390 pKa = 11.84YY391 pKa = 7.63TEE393 pKa = 4.53YY394 pKa = 10.54KK395 pKa = 9.43SQMNRR400 pKa = 11.84VAGDD404 pKa = 3.33FRR406 pKa = 11.84TSLDD410 pKa = 3.3YY411 pKa = 10.5WHH413 pKa = 7.71LARR416 pKa = 11.84KK417 pKa = 8.18FASLPALAQEE427 pKa = 4.58FLEE430 pKa = 4.53ISQGADD436 pKa = 2.82MDD438 pKa = 4.97RR439 pKa = 11.84IFAVIDD445 pKa = 3.32PTVQHH450 pKa = 6.35VLVHH454 pKa = 6.47WFHH457 pKa = 7.14AIKK460 pKa = 10.31VHH462 pKa = 6.35RR463 pKa = 11.84ALPRR467 pKa = 11.84IVTPDD472 pKa = 3.13LL473 pKa = 3.94
Molecular weight: 52.96 kDa
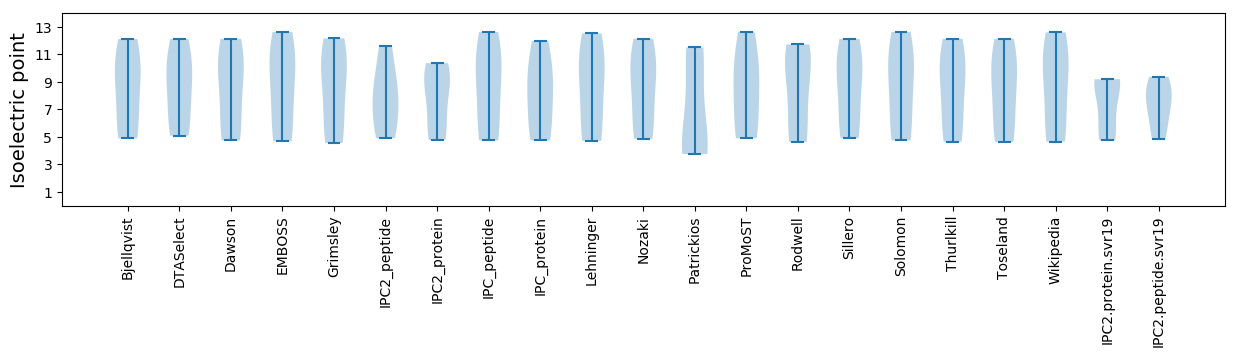
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UG46|A0A0G2UG46_9VIRU Uncharacterized protein OS=Microviridae Fen685_11 OX=1655657 PE=4 SV=1
MM1 pKa = 7.57IFFIVLIVVTSSSLRR16 pKa = 11.84TALLIGLQSLFRR28 pKa = 11.84VSLLLSVIFLLAFLVDD44 pKa = 4.64KK45 pKa = 11.23LFLQLPHH52 pKa = 7.49RR53 pKa = 11.84LRR55 pKa = 11.84ILMLTCPSLIIWIPLTVLLLLVVIVSALRR84 pKa = 11.84LWNRR88 pKa = 11.84RR89 pKa = 3.35
MM1 pKa = 7.57IFFIVLIVVTSSSLRR16 pKa = 11.84TALLIGLQSLFRR28 pKa = 11.84VSLLLSVIFLLAFLVDD44 pKa = 4.64KK45 pKa = 11.23LFLQLPHH52 pKa = 7.49RR53 pKa = 11.84LRR55 pKa = 11.84ILMLTCPSLIIWIPLTVLLLLVVIVSALRR84 pKa = 11.84LWNRR88 pKa = 11.84RR89 pKa = 3.35
Molecular weight: 10.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322 |
89 |
473 |
264.4 |
29.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.985 ± 2.404 | 1.059 ± 0.747 |
6.203 ± 0.836 | 3.328 ± 0.57 |
4.085 ± 0.668 | 5.295 ± 1.196 |
2.269 ± 0.74 | 4.917 ± 0.742 |
3.858 ± 1.106 | 9.834 ± 2.091 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.345 ± 0.304 | 4.766 ± 1.5 |
4.992 ± 0.674 | 5.446 ± 1.186 |
6.278 ± 1.361 | 7.035 ± 0.602 |
5.976 ± 0.489 | 6.43 ± 0.456 |
2.118 ± 0.658 | 3.782 ± 0.768 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
