
Capybara microvirus Cap3_SP_344
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
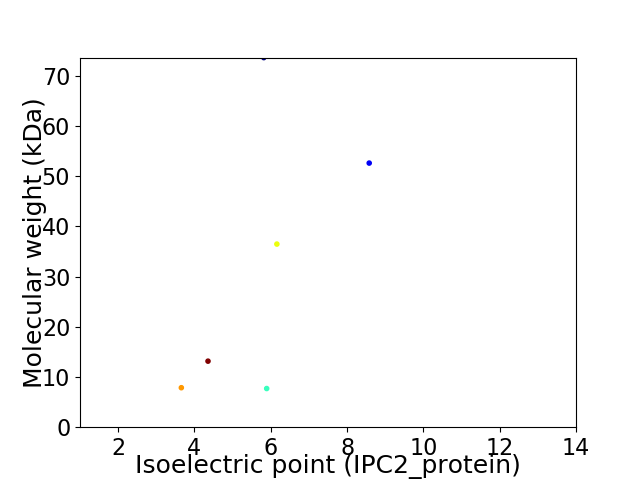
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
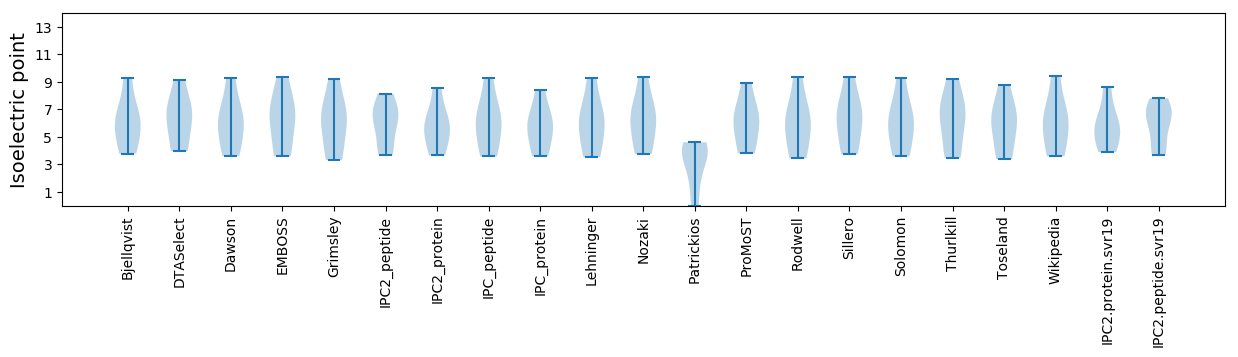
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8E2|A0A4P8W8E2_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_344 OX=2585432 PE=4 SV=1
MM1 pKa = 7.22HH2 pKa = 7.18NCWPSTPCGLVALTGDD18 pKa = 3.17ITQCEE23 pKa = 4.48DD24 pKa = 4.27CPDD27 pKa = 3.24IDD29 pKa = 5.37ARR31 pKa = 11.84FADD34 pKa = 4.07SFFEE38 pKa = 3.92EE39 pKa = 4.58TPRR42 pKa = 11.84YY43 pKa = 9.87DD44 pKa = 3.66EE45 pKa = 4.97RR46 pKa = 11.84CALMNDD52 pKa = 3.03ICSFSYY58 pKa = 10.8VDD60 pKa = 3.76EE61 pKa = 4.97NGDD64 pKa = 3.37YY65 pKa = 11.07DD66 pKa = 5.11LPFF69 pKa = 5.49
MM1 pKa = 7.22HH2 pKa = 7.18NCWPSTPCGLVALTGDD18 pKa = 3.17ITQCEE23 pKa = 4.48DD24 pKa = 4.27CPDD27 pKa = 3.24IDD29 pKa = 5.37ARR31 pKa = 11.84FADD34 pKa = 4.07SFFEE38 pKa = 3.92EE39 pKa = 4.58TPRR42 pKa = 11.84YY43 pKa = 9.87DD44 pKa = 3.66EE45 pKa = 4.97RR46 pKa = 11.84CALMNDD52 pKa = 3.03ICSFSYY58 pKa = 10.8VDD60 pKa = 3.76EE61 pKa = 4.97NGDD64 pKa = 3.37YY65 pKa = 11.07DD66 pKa = 5.11LPFF69 pKa = 5.49
Molecular weight: 7.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4T0|A0A4P8W4T0_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_344 OX=2585432 PE=4 SV=1
MM1 pKa = 7.65CLFPKK6 pKa = 10.49RR7 pKa = 11.84IRR9 pKa = 11.84NPKK12 pKa = 8.67KK13 pKa = 10.26DD14 pKa = 3.53FRR16 pKa = 11.84SGYY19 pKa = 9.65DD20 pKa = 3.74PEE22 pKa = 4.59WLTVPCGNCYY32 pKa = 10.86DD33 pKa = 5.17CIQKK37 pKa = 10.59NIRR40 pKa = 11.84DD41 pKa = 3.31WSARR45 pKa = 11.84ARR47 pKa = 11.84AEE49 pKa = 3.73YY50 pKa = 10.24HH51 pKa = 6.95DD52 pKa = 4.27YY53 pKa = 10.86KK54 pKa = 11.25EE55 pKa = 4.46KK56 pKa = 10.73GGCVLFPTLTYY67 pKa = 10.72NEE69 pKa = 4.45KK70 pKa = 10.7CCPCVNIGNKK80 pKa = 8.76SYY82 pKa = 10.98RR83 pKa = 11.84AFSKK87 pKa = 10.6RR88 pKa = 11.84DD89 pKa = 3.24VQHH92 pKa = 6.7FIKK95 pKa = 10.54KK96 pKa = 8.82FRR98 pKa = 11.84VYY100 pKa = 10.65VSRR103 pKa = 11.84TYY105 pKa = 10.96GKK107 pKa = 10.24EE108 pKa = 3.36FARR111 pKa = 11.84GIKK114 pKa = 10.43YY115 pKa = 9.45MIASEE120 pKa = 4.52FGTSSGKK127 pKa = 6.23THH129 pKa = 6.94RR130 pKa = 11.84PHH132 pKa = 5.38YY133 pKa = 10.21HH134 pKa = 6.75CLLFLPWRR142 pKa = 11.84PKK144 pKa = 9.39IRR146 pKa = 11.84KK147 pKa = 8.01ISSIIRR153 pKa = 11.84KK154 pKa = 7.33SWRR157 pKa = 11.84FGYY160 pKa = 9.63MSWSKK165 pKa = 11.15KK166 pKa = 9.35HH167 pKa = 5.37GAVINSVKK175 pKa = 10.49GISYY179 pKa = 9.0VCKK182 pKa = 10.64YY183 pKa = 8.65MCKK186 pKa = 9.8QSNWYY191 pKa = 9.51HH192 pKa = 4.87FTGDD196 pKa = 3.44VPRR199 pKa = 11.84YY200 pKa = 9.06IYY202 pKa = 10.82KK203 pKa = 10.64GDD205 pKa = 3.86DD206 pKa = 3.45RR207 pKa = 11.84YY208 pKa = 11.39CNPDD212 pKa = 3.21FEE214 pKa = 5.16KK215 pKa = 10.78LSKK218 pKa = 10.72VKK220 pKa = 10.1PFHH223 pKa = 6.03LQSIGFGLCLNKK235 pKa = 10.41YY236 pKa = 9.68VDD238 pKa = 3.46TSEE241 pKa = 5.34IIDD244 pKa = 3.78FRR246 pKa = 11.84FHH248 pKa = 6.58FPKK251 pKa = 10.7DD252 pKa = 3.25SAVPDD257 pKa = 4.01DD258 pKa = 3.63SCYY261 pKa = 9.94YY262 pKa = 9.88TIPQYY267 pKa = 10.98NLHH270 pKa = 6.38YY271 pKa = 10.34LIQDD275 pKa = 4.05KK276 pKa = 10.83IDD278 pKa = 4.13GEE280 pKa = 3.99WSYY283 pKa = 12.13KK284 pKa = 9.95NKK286 pKa = 10.21TYY288 pKa = 11.47DD289 pKa = 3.28FMQKK293 pKa = 9.66VFDD296 pKa = 4.46FKK298 pKa = 11.4VRR300 pKa = 11.84CNWYY304 pKa = 9.85HH305 pKa = 7.19LACQLDD311 pKa = 3.88KK312 pKa = 11.51DD313 pKa = 4.59LLNSCGISEE322 pKa = 4.35FNKK325 pKa = 10.71KK326 pKa = 9.04IDD328 pKa = 3.89NIFDD332 pKa = 3.79EE333 pKa = 4.56TSLNYY338 pKa = 10.11RR339 pKa = 11.84EE340 pKa = 4.46IANFMVSYY348 pKa = 10.56FGTNYY353 pKa = 9.54FPPIDD358 pKa = 4.61FYY360 pKa = 11.79CDD362 pKa = 3.18LGSSFRR368 pKa = 11.84HH369 pKa = 5.21WYY371 pKa = 8.29MRR373 pKa = 11.84YY374 pKa = 9.95LDD376 pKa = 4.41FDD378 pKa = 4.42SDD380 pKa = 3.69SRR382 pKa = 11.84RR383 pKa = 11.84AKK385 pKa = 10.11SDD387 pKa = 3.36CVTYY391 pKa = 9.88EE392 pKa = 4.01YY393 pKa = 11.18KK394 pKa = 10.78KK395 pKa = 9.48NTPLCYY401 pKa = 10.26VSLLLDD407 pKa = 4.06SIRR410 pKa = 11.84LSVEE414 pKa = 3.78NYY416 pKa = 10.15VKK418 pKa = 10.2GTTDD422 pKa = 3.77LKK424 pKa = 9.72RR425 pKa = 11.84HH426 pKa = 5.61KK427 pKa = 9.9FASRR431 pKa = 11.84KK432 pKa = 9.58KK433 pKa = 10.18VIDD436 pKa = 3.59YY437 pKa = 10.76NSNFSS442 pKa = 3.4
MM1 pKa = 7.65CLFPKK6 pKa = 10.49RR7 pKa = 11.84IRR9 pKa = 11.84NPKK12 pKa = 8.67KK13 pKa = 10.26DD14 pKa = 3.53FRR16 pKa = 11.84SGYY19 pKa = 9.65DD20 pKa = 3.74PEE22 pKa = 4.59WLTVPCGNCYY32 pKa = 10.86DD33 pKa = 5.17CIQKK37 pKa = 10.59NIRR40 pKa = 11.84DD41 pKa = 3.31WSARR45 pKa = 11.84ARR47 pKa = 11.84AEE49 pKa = 3.73YY50 pKa = 10.24HH51 pKa = 6.95DD52 pKa = 4.27YY53 pKa = 10.86KK54 pKa = 11.25EE55 pKa = 4.46KK56 pKa = 10.73GGCVLFPTLTYY67 pKa = 10.72NEE69 pKa = 4.45KK70 pKa = 10.7CCPCVNIGNKK80 pKa = 8.76SYY82 pKa = 10.98RR83 pKa = 11.84AFSKK87 pKa = 10.6RR88 pKa = 11.84DD89 pKa = 3.24VQHH92 pKa = 6.7FIKK95 pKa = 10.54KK96 pKa = 8.82FRR98 pKa = 11.84VYY100 pKa = 10.65VSRR103 pKa = 11.84TYY105 pKa = 10.96GKK107 pKa = 10.24EE108 pKa = 3.36FARR111 pKa = 11.84GIKK114 pKa = 10.43YY115 pKa = 9.45MIASEE120 pKa = 4.52FGTSSGKK127 pKa = 6.23THH129 pKa = 6.94RR130 pKa = 11.84PHH132 pKa = 5.38YY133 pKa = 10.21HH134 pKa = 6.75CLLFLPWRR142 pKa = 11.84PKK144 pKa = 9.39IRR146 pKa = 11.84KK147 pKa = 8.01ISSIIRR153 pKa = 11.84KK154 pKa = 7.33SWRR157 pKa = 11.84FGYY160 pKa = 9.63MSWSKK165 pKa = 11.15KK166 pKa = 9.35HH167 pKa = 5.37GAVINSVKK175 pKa = 10.49GISYY179 pKa = 9.0VCKK182 pKa = 10.64YY183 pKa = 8.65MCKK186 pKa = 9.8QSNWYY191 pKa = 9.51HH192 pKa = 4.87FTGDD196 pKa = 3.44VPRR199 pKa = 11.84YY200 pKa = 9.06IYY202 pKa = 10.82KK203 pKa = 10.64GDD205 pKa = 3.86DD206 pKa = 3.45RR207 pKa = 11.84YY208 pKa = 11.39CNPDD212 pKa = 3.21FEE214 pKa = 5.16KK215 pKa = 10.78LSKK218 pKa = 10.72VKK220 pKa = 10.1PFHH223 pKa = 6.03LQSIGFGLCLNKK235 pKa = 10.41YY236 pKa = 9.68VDD238 pKa = 3.46TSEE241 pKa = 5.34IIDD244 pKa = 3.78FRR246 pKa = 11.84FHH248 pKa = 6.58FPKK251 pKa = 10.7DD252 pKa = 3.25SAVPDD257 pKa = 4.01DD258 pKa = 3.63SCYY261 pKa = 9.94YY262 pKa = 9.88TIPQYY267 pKa = 10.98NLHH270 pKa = 6.38YY271 pKa = 10.34LIQDD275 pKa = 4.05KK276 pKa = 10.83IDD278 pKa = 4.13GEE280 pKa = 3.99WSYY283 pKa = 12.13KK284 pKa = 9.95NKK286 pKa = 10.21TYY288 pKa = 11.47DD289 pKa = 3.28FMQKK293 pKa = 9.66VFDD296 pKa = 4.46FKK298 pKa = 11.4VRR300 pKa = 11.84CNWYY304 pKa = 9.85HH305 pKa = 7.19LACQLDD311 pKa = 3.88KK312 pKa = 11.51DD313 pKa = 4.59LLNSCGISEE322 pKa = 4.35FNKK325 pKa = 10.71KK326 pKa = 9.04IDD328 pKa = 3.89NIFDD332 pKa = 3.79EE333 pKa = 4.56TSLNYY338 pKa = 10.11RR339 pKa = 11.84EE340 pKa = 4.46IANFMVSYY348 pKa = 10.56FGTNYY353 pKa = 9.54FPPIDD358 pKa = 4.61FYY360 pKa = 11.79CDD362 pKa = 3.18LGSSFRR368 pKa = 11.84HH369 pKa = 5.21WYY371 pKa = 8.29MRR373 pKa = 11.84YY374 pKa = 9.95LDD376 pKa = 4.41FDD378 pKa = 4.42SDD380 pKa = 3.69SRR382 pKa = 11.84RR383 pKa = 11.84AKK385 pKa = 10.11SDD387 pKa = 3.36CVTYY391 pKa = 9.88EE392 pKa = 4.01YY393 pKa = 11.18KK394 pKa = 10.78KK395 pKa = 9.48NTPLCYY401 pKa = 10.26VSLLLDD407 pKa = 4.06SIRR410 pKa = 11.84LSVEE414 pKa = 3.78NYY416 pKa = 10.15VKK418 pKa = 10.2GTTDD422 pKa = 3.77LKK424 pKa = 9.72RR425 pKa = 11.84HH426 pKa = 5.61KK427 pKa = 9.9FASRR431 pKa = 11.84KK432 pKa = 9.58KK433 pKa = 10.18VIDD436 pKa = 3.59YY437 pKa = 10.76NSNFSS442 pKa = 3.4
Molecular weight: 52.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1693 |
67 |
653 |
282.2 |
31.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.675 ± 1.868 | 2.185 ± 0.966 |
5.848 ± 0.963 | 5.08 ± 1.131 |
5.552 ± 0.942 | 6.084 ± 1.001 |
1.89 ± 0.428 | 4.607 ± 0.487 |
6.261 ± 1.152 | 8.328 ± 1.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.067 ± 0.324 | 5.611 ± 0.308 |
4.017 ± 0.375 | 3.721 ± 0.722 |
4.371 ± 0.671 | 9.569 ± 0.644 |
4.903 ± 0.834 | 6.143 ± 0.585 |
1.24 ± 0.431 | 5.848 ± 1.252 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
