
Hubei sobemo-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
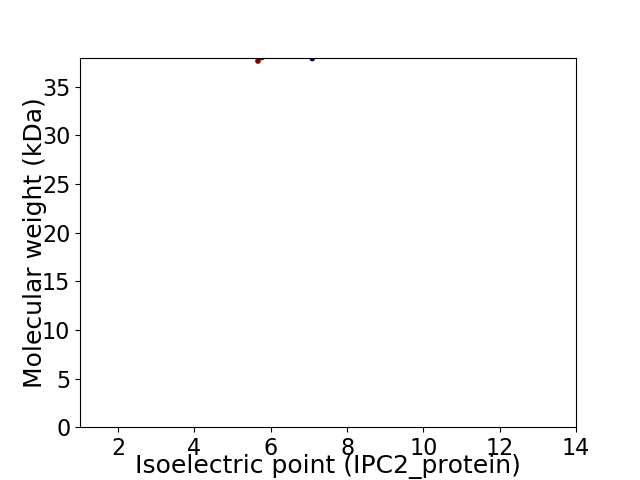
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KER3|A0A1L3KER3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 12 OX=1923197 PE=4 SV=1
MM1 pKa = 7.95DD2 pKa = 4.98FDD4 pKa = 3.64QARR7 pKa = 11.84VQDD10 pKa = 3.38LHH12 pKa = 8.34AVVFEE17 pKa = 5.06RR18 pKa = 11.84IRR20 pKa = 11.84QLEE23 pKa = 4.07AGEE26 pKa = 4.35YY27 pKa = 8.95VCDD30 pKa = 4.68PIKK33 pKa = 11.11VFVKK37 pKa = 10.06QEE39 pKa = 3.48PHH41 pKa = 6.22KK42 pKa = 10.92AKK44 pKa = 10.69KK45 pKa = 8.36LTNEE49 pKa = 3.96KK50 pKa = 10.09YY51 pKa = 10.1RR52 pKa = 11.84LISAVSLVDD61 pKa = 3.22TMIDD65 pKa = 3.59RR66 pKa = 11.84ILGRR70 pKa = 11.84SFFEE74 pKa = 4.95SVLEE78 pKa = 4.19NNLEE82 pKa = 4.15VPSAIGWSPNKK93 pKa = 10.06GGYY96 pKa = 8.79RR97 pKa = 11.84ALRR100 pKa = 11.84AIFPKK105 pKa = 10.36GEE107 pKa = 4.09KK108 pKa = 10.5VFTSDD113 pKa = 4.49KK114 pKa = 11.39SNWDD118 pKa = 2.92WTVQKK123 pKa = 10.4WLICAFFEE131 pKa = 5.05VVNEE135 pKa = 4.19LCYY138 pKa = 10.84TEE140 pKa = 5.89DD141 pKa = 3.5KK142 pKa = 10.8EE143 pKa = 5.78LYY145 pKa = 8.42QRR147 pKa = 11.84WYY149 pKa = 9.01ALLWKK154 pKa = 10.27RR155 pKa = 11.84CEE157 pKa = 4.04LLFNRR162 pKa = 11.84AFFKK166 pKa = 10.98FQDD169 pKa = 3.74GTIVQQKK176 pKa = 10.58LPGLQKK182 pKa = 10.52SGCYY186 pKa = 6.46WTIVLNTVGQDD197 pKa = 3.25ILHH200 pKa = 5.81VAAGGVGRR208 pKa = 11.84ILGLGDD214 pKa = 5.67DD215 pKa = 3.93IAQEE219 pKa = 4.29VEE221 pKa = 4.04VDD223 pKa = 3.35PEE225 pKa = 4.19KK226 pKa = 10.94YY227 pKa = 10.03LEE229 pKa = 4.0RR230 pKa = 11.84VQRR233 pKa = 11.84YY234 pKa = 7.55GCEE237 pKa = 3.84VKK239 pKa = 10.47EE240 pKa = 3.97FSIDD244 pKa = 3.31KK245 pKa = 10.6VFDD248 pKa = 3.63FGGYY252 pKa = 9.38HH253 pKa = 5.85YY254 pKa = 10.06EE255 pKa = 4.35GEE257 pKa = 4.34IPTPVYY263 pKa = 10.04HH264 pKa = 6.69GKK266 pKa = 9.26HH267 pKa = 5.48VYY269 pKa = 10.41QMVHH273 pKa = 7.31AEE275 pKa = 4.15VGDD278 pKa = 3.75VALWDD283 pKa = 4.1SYY285 pKa = 11.2LGNYY289 pKa = 9.66AGDD292 pKa = 3.97EE293 pKa = 4.18EE294 pKa = 4.75WFEE297 pKa = 4.88AIRR300 pKa = 11.84KK301 pKa = 5.57VARR304 pKa = 11.84RR305 pKa = 11.84VTGRR309 pKa = 11.84IPKK312 pKa = 9.43SRR314 pKa = 11.84NYY316 pKa = 10.35YY317 pKa = 9.61LSLWKK322 pKa = 10.75DD323 pKa = 3.37EE324 pKa = 4.23
MM1 pKa = 7.95DD2 pKa = 4.98FDD4 pKa = 3.64QARR7 pKa = 11.84VQDD10 pKa = 3.38LHH12 pKa = 8.34AVVFEE17 pKa = 5.06RR18 pKa = 11.84IRR20 pKa = 11.84QLEE23 pKa = 4.07AGEE26 pKa = 4.35YY27 pKa = 8.95VCDD30 pKa = 4.68PIKK33 pKa = 11.11VFVKK37 pKa = 10.06QEE39 pKa = 3.48PHH41 pKa = 6.22KK42 pKa = 10.92AKK44 pKa = 10.69KK45 pKa = 8.36LTNEE49 pKa = 3.96KK50 pKa = 10.09YY51 pKa = 10.1RR52 pKa = 11.84LISAVSLVDD61 pKa = 3.22TMIDD65 pKa = 3.59RR66 pKa = 11.84ILGRR70 pKa = 11.84SFFEE74 pKa = 4.95SVLEE78 pKa = 4.19NNLEE82 pKa = 4.15VPSAIGWSPNKK93 pKa = 10.06GGYY96 pKa = 8.79RR97 pKa = 11.84ALRR100 pKa = 11.84AIFPKK105 pKa = 10.36GEE107 pKa = 4.09KK108 pKa = 10.5VFTSDD113 pKa = 4.49KK114 pKa = 11.39SNWDD118 pKa = 2.92WTVQKK123 pKa = 10.4WLICAFFEE131 pKa = 5.05VVNEE135 pKa = 4.19LCYY138 pKa = 10.84TEE140 pKa = 5.89DD141 pKa = 3.5KK142 pKa = 10.8EE143 pKa = 5.78LYY145 pKa = 8.42QRR147 pKa = 11.84WYY149 pKa = 9.01ALLWKK154 pKa = 10.27RR155 pKa = 11.84CEE157 pKa = 4.04LLFNRR162 pKa = 11.84AFFKK166 pKa = 10.98FQDD169 pKa = 3.74GTIVQQKK176 pKa = 10.58LPGLQKK182 pKa = 10.52SGCYY186 pKa = 6.46WTIVLNTVGQDD197 pKa = 3.25ILHH200 pKa = 5.81VAAGGVGRR208 pKa = 11.84ILGLGDD214 pKa = 5.67DD215 pKa = 3.93IAQEE219 pKa = 4.29VEE221 pKa = 4.04VDD223 pKa = 3.35PEE225 pKa = 4.19KK226 pKa = 10.94YY227 pKa = 10.03LEE229 pKa = 4.0RR230 pKa = 11.84VQRR233 pKa = 11.84YY234 pKa = 7.55GCEE237 pKa = 3.84VKK239 pKa = 10.47EE240 pKa = 3.97FSIDD244 pKa = 3.31KK245 pKa = 10.6VFDD248 pKa = 3.63FGGYY252 pKa = 9.38HH253 pKa = 5.85YY254 pKa = 10.06EE255 pKa = 4.35GEE257 pKa = 4.34IPTPVYY263 pKa = 10.04HH264 pKa = 6.69GKK266 pKa = 9.26HH267 pKa = 5.48VYY269 pKa = 10.41QMVHH273 pKa = 7.31AEE275 pKa = 4.15VGDD278 pKa = 3.75VALWDD283 pKa = 4.1SYY285 pKa = 11.2LGNYY289 pKa = 9.66AGDD292 pKa = 3.97EE293 pKa = 4.18EE294 pKa = 4.75WFEE297 pKa = 4.88AIRR300 pKa = 11.84KK301 pKa = 5.57VARR304 pKa = 11.84RR305 pKa = 11.84VTGRR309 pKa = 11.84IPKK312 pKa = 9.43SRR314 pKa = 11.84NYY316 pKa = 10.35YY317 pKa = 9.61LSLWKK322 pKa = 10.75DD323 pKa = 3.37EE324 pKa = 4.23
Molecular weight: 37.65 kDa
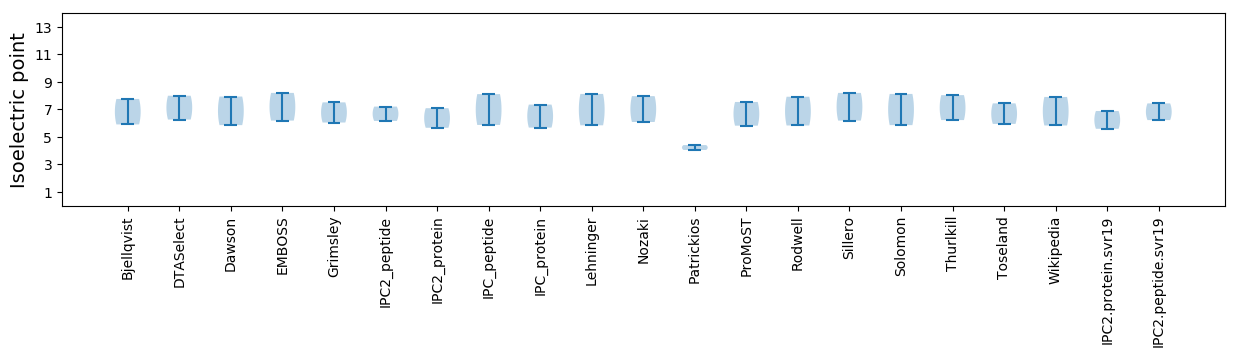
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KER3|A0A1L3KER3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 12 OX=1923197 PE=4 SV=1
MM1 pKa = 7.67GFRR4 pKa = 11.84MDD6 pKa = 3.91NNLITTLHH14 pKa = 6.07VIDD17 pKa = 5.12RR18 pKa = 11.84LCEE21 pKa = 3.82IQIKK25 pKa = 7.13TDD27 pKa = 2.84KK28 pKa = 10.28GAVMVHH34 pKa = 6.11KK35 pKa = 10.82EE36 pKa = 3.91EE37 pKa = 4.15FQEE40 pKa = 4.1LDD42 pKa = 3.54NEE44 pKa = 4.86VVFAPYY50 pKa = 7.99TQNMQVLGLAKK61 pKa = 10.46YY62 pKa = 10.13KK63 pKa = 10.31IATVGMSMFDD73 pKa = 3.33MAVVRR78 pKa = 11.84IVGKK82 pKa = 8.98MPYY85 pKa = 9.96VRR87 pKa = 11.84TSGLLKK93 pKa = 10.31KK94 pKa = 9.7NRR96 pKa = 11.84SMFYY100 pKa = 10.36VSYY103 pKa = 10.5HH104 pKa = 6.03GSTAKK109 pKa = 10.43GFSGAPYY116 pKa = 8.43VQGRR120 pKa = 11.84NIYY123 pKa = 8.58GMHH126 pKa = 6.4MGSTTGTHH134 pKa = 6.38NMGLDD139 pKa = 3.27ATYY142 pKa = 9.62IQAMVRR148 pKa = 11.84GLDD151 pKa = 3.43EE152 pKa = 4.72AYY154 pKa = 9.55PWEE157 pKa = 4.6KK158 pKa = 10.09YY159 pKa = 9.63KK160 pKa = 10.87KK161 pKa = 9.38RR162 pKa = 11.84AARR165 pKa = 11.84MKK167 pKa = 10.88GKK169 pKa = 8.25VTPSGGYY176 pKa = 10.37LEE178 pKa = 4.49THH180 pKa = 7.03DD181 pKa = 3.87GYY183 pKa = 11.77KK184 pKa = 10.16RR185 pKa = 11.84VDD187 pKa = 3.0ASKK190 pKa = 11.35YY191 pKa = 9.96NDD193 pKa = 3.61LVEE196 pKa = 4.25QGIVDD201 pKa = 3.48WGEE204 pKa = 4.25LMDD207 pKa = 5.21KK208 pKa = 10.21MEE210 pKa = 4.45YY211 pKa = 10.09DD212 pKa = 3.4QPEE215 pKa = 4.12ADD217 pKa = 3.91LPSANRR223 pKa = 11.84EE224 pKa = 4.13HH225 pKa = 7.22LEE227 pKa = 4.32HH228 pKa = 7.67DD229 pKa = 5.11DD230 pKa = 4.26SDD232 pKa = 3.99SDD234 pKa = 3.31QDD236 pKa = 3.49FRR238 pKa = 11.84SRR240 pKa = 11.84GLPTMDD246 pKa = 3.72RR247 pKa = 11.84PRR249 pKa = 11.84AVASNCPIAGISTDD263 pKa = 3.29PAEE266 pKa = 4.28VVFYY270 pKa = 10.39PGSQQKK276 pKa = 10.09QGTPILTNQTFQHH289 pKa = 5.7SRR291 pKa = 11.84EE292 pKa = 4.17FTDD295 pKa = 3.15HH296 pKa = 7.04LEE298 pKa = 4.5SILAQRR304 pKa = 11.84NDD306 pKa = 3.57LSPTIRR312 pKa = 11.84RR313 pKa = 11.84SLRR316 pKa = 11.84KK317 pKa = 8.47ATNFLKK323 pKa = 10.57SQAMKK328 pKa = 10.94NSTEE332 pKa = 4.03SSKK335 pKa = 10.33QQ336 pKa = 3.26
MM1 pKa = 7.67GFRR4 pKa = 11.84MDD6 pKa = 3.91NNLITTLHH14 pKa = 6.07VIDD17 pKa = 5.12RR18 pKa = 11.84LCEE21 pKa = 3.82IQIKK25 pKa = 7.13TDD27 pKa = 2.84KK28 pKa = 10.28GAVMVHH34 pKa = 6.11KK35 pKa = 10.82EE36 pKa = 3.91EE37 pKa = 4.15FQEE40 pKa = 4.1LDD42 pKa = 3.54NEE44 pKa = 4.86VVFAPYY50 pKa = 7.99TQNMQVLGLAKK61 pKa = 10.46YY62 pKa = 10.13KK63 pKa = 10.31IATVGMSMFDD73 pKa = 3.33MAVVRR78 pKa = 11.84IVGKK82 pKa = 8.98MPYY85 pKa = 9.96VRR87 pKa = 11.84TSGLLKK93 pKa = 10.31KK94 pKa = 9.7NRR96 pKa = 11.84SMFYY100 pKa = 10.36VSYY103 pKa = 10.5HH104 pKa = 6.03GSTAKK109 pKa = 10.43GFSGAPYY116 pKa = 8.43VQGRR120 pKa = 11.84NIYY123 pKa = 8.58GMHH126 pKa = 6.4MGSTTGTHH134 pKa = 6.38NMGLDD139 pKa = 3.27ATYY142 pKa = 9.62IQAMVRR148 pKa = 11.84GLDD151 pKa = 3.43EE152 pKa = 4.72AYY154 pKa = 9.55PWEE157 pKa = 4.6KK158 pKa = 10.09YY159 pKa = 9.63KK160 pKa = 10.87KK161 pKa = 9.38RR162 pKa = 11.84AARR165 pKa = 11.84MKK167 pKa = 10.88GKK169 pKa = 8.25VTPSGGYY176 pKa = 10.37LEE178 pKa = 4.49THH180 pKa = 7.03DD181 pKa = 3.87GYY183 pKa = 11.77KK184 pKa = 10.16RR185 pKa = 11.84VDD187 pKa = 3.0ASKK190 pKa = 11.35YY191 pKa = 9.96NDD193 pKa = 3.61LVEE196 pKa = 4.25QGIVDD201 pKa = 3.48WGEE204 pKa = 4.25LMDD207 pKa = 5.21KK208 pKa = 10.21MEE210 pKa = 4.45YY211 pKa = 10.09DD212 pKa = 3.4QPEE215 pKa = 4.12ADD217 pKa = 3.91LPSANRR223 pKa = 11.84EE224 pKa = 4.13HH225 pKa = 7.22LEE227 pKa = 4.32HH228 pKa = 7.67DD229 pKa = 5.11DD230 pKa = 4.26SDD232 pKa = 3.99SDD234 pKa = 3.31QDD236 pKa = 3.49FRR238 pKa = 11.84SRR240 pKa = 11.84GLPTMDD246 pKa = 3.72RR247 pKa = 11.84PRR249 pKa = 11.84AVASNCPIAGISTDD263 pKa = 3.29PAEE266 pKa = 4.28VVFYY270 pKa = 10.39PGSQQKK276 pKa = 10.09QGTPILTNQTFQHH289 pKa = 5.7SRR291 pKa = 11.84EE292 pKa = 4.17FTDD295 pKa = 3.15HH296 pKa = 7.04LEE298 pKa = 4.5SILAQRR304 pKa = 11.84NDD306 pKa = 3.57LSPTIRR312 pKa = 11.84RR313 pKa = 11.84SLRR316 pKa = 11.84KK317 pKa = 8.47ATNFLKK323 pKa = 10.57SQAMKK328 pKa = 10.94NSTEE332 pKa = 4.03SSKK335 pKa = 10.33QQ336 pKa = 3.26
Molecular weight: 37.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
660 |
324 |
336 |
330.0 |
37.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.212 ± 0.238 | 1.212 ± 0.438 |
6.515 ± 0.234 | 6.818 ± 1.038 |
4.242 ± 0.688 | 7.424 ± 0.012 |
2.576 ± 0.284 | 4.697 ± 0.377 |
6.818 ± 0.192 | 7.424 ± 0.623 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.182 ± 1.545 | 3.636 ± 0.377 |
3.636 ± 0.377 | 4.697 ± 0.258 |
5.909 ± 0.031 | 5.606 ± 1.092 |
4.848 ± 1.207 | 7.879 ± 1.157 |
1.818 ± 0.869 | 4.848 ± 0.273 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
