
Capybara microvirus Cap1_SP_55
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
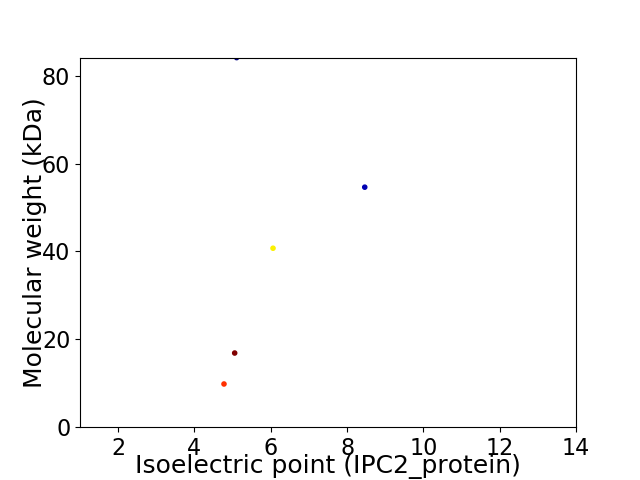
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4N1|A0A4P8W4N1_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_55 OX=2584783 PE=4 SV=1
MM1 pKa = 7.1FRR3 pKa = 11.84CRR5 pKa = 11.84QPIRR9 pKa = 11.84FSPSSSISLIEE20 pKa = 3.98PSLVDD25 pKa = 3.8DD26 pKa = 4.6VYY28 pKa = 11.09VCKK31 pKa = 10.51EE32 pKa = 3.23IDD34 pKa = 3.19QCRR37 pKa = 11.84SLPSSDD43 pKa = 4.54NMDD46 pKa = 4.07LKK48 pKa = 10.4TLLDD52 pKa = 4.1AGLPIDD58 pKa = 4.2EE59 pKa = 4.69VSTIIRR65 pKa = 11.84SGSVDD70 pKa = 3.58LSEE73 pKa = 4.27LAKK76 pKa = 10.62KK77 pKa = 10.72YY78 pKa = 10.58GVEE81 pKa = 3.89IKK83 pKa = 10.84NKK85 pKa = 9.61EE86 pKa = 3.89NEE88 pKa = 3.94
MM1 pKa = 7.1FRR3 pKa = 11.84CRR5 pKa = 11.84QPIRR9 pKa = 11.84FSPSSSISLIEE20 pKa = 3.98PSLVDD25 pKa = 3.8DD26 pKa = 4.6VYY28 pKa = 11.09VCKK31 pKa = 10.51EE32 pKa = 3.23IDD34 pKa = 3.19QCRR37 pKa = 11.84SLPSSDD43 pKa = 4.54NMDD46 pKa = 4.07LKK48 pKa = 10.4TLLDD52 pKa = 4.1AGLPIDD58 pKa = 4.2EE59 pKa = 4.69VSTIIRR65 pKa = 11.84SGSVDD70 pKa = 3.58LSEE73 pKa = 4.27LAKK76 pKa = 10.62KK77 pKa = 10.72YY78 pKa = 10.58GVEE81 pKa = 3.89IKK83 pKa = 10.84NKK85 pKa = 9.61EE86 pKa = 3.89NEE88 pKa = 3.94
Molecular weight: 9.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7G1|A0A4P8W7G1_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_55 OX=2584783 PE=4 SV=1
MM1 pKa = 7.41NLNNFRR7 pKa = 11.84VKK9 pKa = 10.39SISPCSPKK17 pKa = 10.38KK18 pKa = 10.86VFVSNPRR25 pKa = 11.84SYY27 pKa = 11.62SSIKK31 pKa = 10.4AKK33 pKa = 10.29SDD35 pKa = 2.87SWSFRR40 pKa = 11.84LAAEE44 pKa = 4.58LKK46 pKa = 9.46NAVEE50 pKa = 4.21NGYY53 pKa = 10.31KK54 pKa = 10.28VGFVTLTYY62 pKa = 11.03NDD64 pKa = 4.4DD65 pKa = 3.61NLPYY69 pKa = 10.57VDD71 pKa = 5.09GSLLRR76 pKa = 11.84DD77 pKa = 3.44VTLDD81 pKa = 3.45SRR83 pKa = 11.84PCFDD87 pKa = 3.78RR88 pKa = 11.84KK89 pKa = 7.9QVRR92 pKa = 11.84LFVDD96 pKa = 5.23NIRR99 pKa = 11.84KK100 pKa = 8.41WLWRR104 pKa = 11.84HH105 pKa = 4.9WRR107 pKa = 11.84ITDD110 pKa = 3.31IKK112 pKa = 9.5YY113 pKa = 7.84TVCSEE118 pKa = 3.88YY119 pKa = 11.39GEE121 pKa = 4.37FTRR124 pKa = 11.84RR125 pKa = 11.84PHH127 pKa = 4.76YY128 pKa = 10.14HH129 pKa = 7.04AVFAWPFKK137 pKa = 10.53AASFYY142 pKa = 10.76IGNDD146 pKa = 3.21EE147 pKa = 4.1NKK149 pKa = 10.31KK150 pKa = 10.52LVTRR154 pKa = 11.84PPFSAAMFHH163 pKa = 7.03ALIKK167 pKa = 10.51RR168 pKa = 11.84FWHH171 pKa = 6.07YY172 pKa = 11.43GFISPRR178 pKa = 11.84DD179 pKa = 3.52YY180 pKa = 11.26RR181 pKa = 11.84GGKK184 pKa = 7.38SCKK187 pKa = 9.65GRR189 pKa = 11.84VYY191 pKa = 10.78EE192 pKa = 4.25PFEE195 pKa = 4.18VDD197 pKa = 3.03PLKK200 pKa = 11.2VVGAACYY207 pKa = 9.58CGKK210 pKa = 10.35YY211 pKa = 9.0CCKK214 pKa = 10.6DD215 pKa = 2.73IAFYY219 pKa = 11.16EE220 pKa = 4.4DD221 pKa = 4.68LPTDD225 pKa = 3.67YY226 pKa = 10.35FVQSDD231 pKa = 3.82DD232 pKa = 3.71VDD234 pKa = 3.68VKK236 pKa = 11.03KK237 pKa = 10.68AIKK240 pKa = 10.3NCMPFHH246 pKa = 6.44LHH248 pKa = 6.2SKK250 pKa = 10.45SLGLCFLDD258 pKa = 5.72KK259 pKa = 10.1ITNEE263 pKa = 3.79DD264 pKa = 3.32KK265 pKa = 11.28LRR267 pKa = 11.84FLSKK271 pKa = 9.64GVQRR275 pKa = 11.84IGSDD279 pKa = 3.01KK280 pKa = 10.52FVSLPIYY287 pKa = 9.68YY288 pKa = 10.04KK289 pKa = 11.0NKK291 pKa = 9.79ILFDD295 pKa = 3.7TYY297 pKa = 11.65YY298 pKa = 11.2SFDD301 pKa = 4.04SNGDD305 pKa = 3.25RR306 pKa = 11.84LVRR309 pKa = 11.84RR310 pKa = 11.84KK311 pKa = 10.01ANKK314 pKa = 9.49FFLDD318 pKa = 3.62NYY320 pKa = 10.37FEE322 pKa = 4.65IYY324 pKa = 9.89HH325 pKa = 6.6IKK327 pKa = 8.75FQAYY331 pKa = 8.95KK332 pKa = 10.64SVFAPLFDD340 pKa = 5.35DD341 pKa = 5.56SYY343 pKa = 11.06LKK345 pKa = 10.94NYY347 pKa = 9.53IDD349 pKa = 5.65DD350 pKa = 4.38VYY352 pKa = 11.52LSDD355 pKa = 3.88LKK357 pKa = 10.87EE358 pKa = 3.85RR359 pKa = 11.84RR360 pKa = 11.84FNLGVGDD367 pKa = 4.35LNSFTMMYY375 pKa = 10.41LSYY378 pKa = 11.43AFVDD382 pKa = 3.95PNFICNDD389 pKa = 3.97EE390 pKa = 4.45IITWLMRR397 pKa = 11.84YY398 pKa = 9.25VPFYY402 pKa = 11.14EE403 pKa = 3.88NYY405 pKa = 8.94KK406 pKa = 10.53FKK408 pKa = 10.85GIKK411 pKa = 9.98FGVSFEE417 pKa = 4.03MKK419 pKa = 9.94TYY421 pKa = 10.52FNNLNRR427 pKa = 11.84LFYY430 pKa = 10.28DD431 pKa = 3.48HH432 pKa = 7.12LVIFCNDD439 pKa = 3.76SISDD443 pKa = 3.71TRR445 pKa = 11.84LDD447 pKa = 4.91DD448 pKa = 3.47YY449 pKa = 11.71RR450 pKa = 11.84KK451 pKa = 6.82TQKK454 pKa = 10.62VKK456 pKa = 10.98DD457 pKa = 4.44FYY459 pKa = 11.58NQQ461 pKa = 2.84
MM1 pKa = 7.41NLNNFRR7 pKa = 11.84VKK9 pKa = 10.39SISPCSPKK17 pKa = 10.38KK18 pKa = 10.86VFVSNPRR25 pKa = 11.84SYY27 pKa = 11.62SSIKK31 pKa = 10.4AKK33 pKa = 10.29SDD35 pKa = 2.87SWSFRR40 pKa = 11.84LAAEE44 pKa = 4.58LKK46 pKa = 9.46NAVEE50 pKa = 4.21NGYY53 pKa = 10.31KK54 pKa = 10.28VGFVTLTYY62 pKa = 11.03NDD64 pKa = 4.4DD65 pKa = 3.61NLPYY69 pKa = 10.57VDD71 pKa = 5.09GSLLRR76 pKa = 11.84DD77 pKa = 3.44VTLDD81 pKa = 3.45SRR83 pKa = 11.84PCFDD87 pKa = 3.78RR88 pKa = 11.84KK89 pKa = 7.9QVRR92 pKa = 11.84LFVDD96 pKa = 5.23NIRR99 pKa = 11.84KK100 pKa = 8.41WLWRR104 pKa = 11.84HH105 pKa = 4.9WRR107 pKa = 11.84ITDD110 pKa = 3.31IKK112 pKa = 9.5YY113 pKa = 7.84TVCSEE118 pKa = 3.88YY119 pKa = 11.39GEE121 pKa = 4.37FTRR124 pKa = 11.84RR125 pKa = 11.84PHH127 pKa = 4.76YY128 pKa = 10.14HH129 pKa = 7.04AVFAWPFKK137 pKa = 10.53AASFYY142 pKa = 10.76IGNDD146 pKa = 3.21EE147 pKa = 4.1NKK149 pKa = 10.31KK150 pKa = 10.52LVTRR154 pKa = 11.84PPFSAAMFHH163 pKa = 7.03ALIKK167 pKa = 10.51RR168 pKa = 11.84FWHH171 pKa = 6.07YY172 pKa = 11.43GFISPRR178 pKa = 11.84DD179 pKa = 3.52YY180 pKa = 11.26RR181 pKa = 11.84GGKK184 pKa = 7.38SCKK187 pKa = 9.65GRR189 pKa = 11.84VYY191 pKa = 10.78EE192 pKa = 4.25PFEE195 pKa = 4.18VDD197 pKa = 3.03PLKK200 pKa = 11.2VVGAACYY207 pKa = 9.58CGKK210 pKa = 10.35YY211 pKa = 9.0CCKK214 pKa = 10.6DD215 pKa = 2.73IAFYY219 pKa = 11.16EE220 pKa = 4.4DD221 pKa = 4.68LPTDD225 pKa = 3.67YY226 pKa = 10.35FVQSDD231 pKa = 3.82DD232 pKa = 3.71VDD234 pKa = 3.68VKK236 pKa = 11.03KK237 pKa = 10.68AIKK240 pKa = 10.3NCMPFHH246 pKa = 6.44LHH248 pKa = 6.2SKK250 pKa = 10.45SLGLCFLDD258 pKa = 5.72KK259 pKa = 10.1ITNEE263 pKa = 3.79DD264 pKa = 3.32KK265 pKa = 11.28LRR267 pKa = 11.84FLSKK271 pKa = 9.64GVQRR275 pKa = 11.84IGSDD279 pKa = 3.01KK280 pKa = 10.52FVSLPIYY287 pKa = 9.68YY288 pKa = 10.04KK289 pKa = 11.0NKK291 pKa = 9.79ILFDD295 pKa = 3.7TYY297 pKa = 11.65YY298 pKa = 11.2SFDD301 pKa = 4.04SNGDD305 pKa = 3.25RR306 pKa = 11.84LVRR309 pKa = 11.84RR310 pKa = 11.84KK311 pKa = 10.01ANKK314 pKa = 9.49FFLDD318 pKa = 3.62NYY320 pKa = 10.37FEE322 pKa = 4.65IYY324 pKa = 9.89HH325 pKa = 6.6IKK327 pKa = 8.75FQAYY331 pKa = 8.95KK332 pKa = 10.64SVFAPLFDD340 pKa = 5.35DD341 pKa = 5.56SYY343 pKa = 11.06LKK345 pKa = 10.94NYY347 pKa = 9.53IDD349 pKa = 5.65DD350 pKa = 4.38VYY352 pKa = 11.52LSDD355 pKa = 3.88LKK357 pKa = 10.87EE358 pKa = 3.85RR359 pKa = 11.84RR360 pKa = 11.84FNLGVGDD367 pKa = 4.35LNSFTMMYY375 pKa = 10.41LSYY378 pKa = 11.43AFVDD382 pKa = 3.95PNFICNDD389 pKa = 3.97EE390 pKa = 4.45IITWLMRR397 pKa = 11.84YY398 pKa = 9.25VPFYY402 pKa = 11.14EE403 pKa = 3.88NYY405 pKa = 8.94KK406 pKa = 10.53FKK408 pKa = 10.85GIKK411 pKa = 9.98FGVSFEE417 pKa = 4.03MKK419 pKa = 9.94TYY421 pKa = 10.52FNNLNRR427 pKa = 11.84LFYY430 pKa = 10.28DD431 pKa = 3.48HH432 pKa = 7.12LVIFCNDD439 pKa = 3.76SISDD443 pKa = 3.71TRR445 pKa = 11.84LDD447 pKa = 4.91DD448 pKa = 3.47YY449 pKa = 11.71RR450 pKa = 11.84KK451 pKa = 6.82TQKK454 pKa = 10.62VKK456 pKa = 10.98DD457 pKa = 4.44FYY459 pKa = 11.58NQQ461 pKa = 2.84
Molecular weight: 54.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1820 |
88 |
751 |
364.0 |
41.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.495 ± 1.265 | 1.429 ± 0.473 |
7.253 ± 0.655 | 4.56 ± 0.846 |
5.824 ± 1.048 | 5.824 ± 0.875 |
1.923 ± 0.143 | 4.945 ± 0.43 |
6.374 ± 0.892 | 9.396 ± 0.737 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.143 ± 0.374 | 5.879 ± 0.359 |
4.341 ± 0.617 | 3.077 ± 0.875 |
4.615 ± 0.549 | 9.286 ± 0.662 |
4.67 ± 0.694 | 6.923 ± 0.474 |
1.264 ± 0.345 | 4.78 ± 0.956 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
