
Lupinus angustifolius (Narrow-leaved blue lupine)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; 50 kb inversion clade; genistoids sensu lato; core genistoids;
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
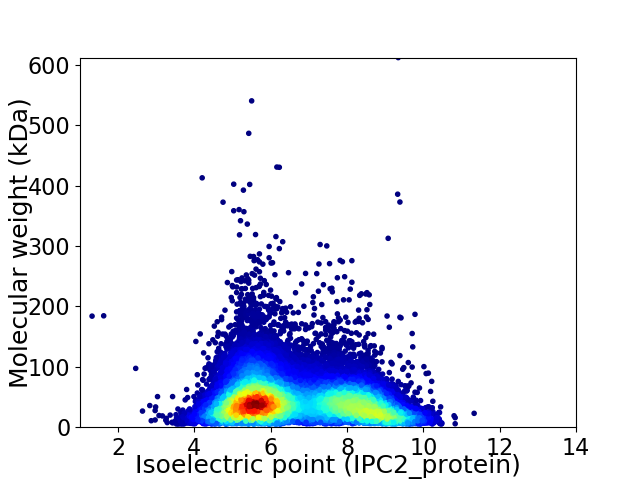
Virtual 2D-PAGE plot for 31106 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J7FMY5|A0A1J7FMY5_LUPAN Glycosyltransferase family 92 protein OS=Lupinus angustifolius OX=3871 GN=TanjilG_23887 PE=3 SV=1
MM1 pKa = 7.76ADD3 pKa = 2.79VTTYY7 pKa = 10.69LRR9 pKa = 11.84HH10 pKa = 6.25HH11 pKa = 6.76KK12 pKa = 9.93PDD14 pKa = 3.57EE15 pKa = 4.68DD16 pKa = 4.19FDD18 pKa = 6.28SITLPYY24 pKa = 9.97FSQSSPDD31 pKa = 3.59FNFDD35 pKa = 3.88FYY37 pKa = 11.45SSEE40 pKa = 4.31PEE42 pKa = 4.01FPPTPEE48 pKa = 4.94PDD50 pKa = 3.49SLSLFDD56 pKa = 4.57RR57 pKa = 11.84HH58 pKa = 5.39NQINFAIDD66 pKa = 3.54IFQQCVDD73 pKa = 3.39QSQVSGHH80 pKa = 6.3GYY82 pKa = 9.93FVTEE86 pKa = 4.54PRR88 pKa = 11.84NDD90 pKa = 3.74DD91 pKa = 3.63DD92 pKa = 5.43FGVIDD97 pKa = 4.58VYY99 pKa = 11.45SALDD103 pKa = 3.34MDD105 pKa = 5.88DD106 pKa = 5.14NLGLDD111 pKa = 3.45LGFGVEE117 pKa = 4.18RR118 pKa = 11.84DD119 pKa = 3.55GVSISEE125 pKa = 4.22SNEE128 pKa = 3.81APFHH132 pKa = 5.92NCVCVMGLGSDD143 pKa = 3.42SDD145 pKa = 3.89EE146 pKa = 4.1EE147 pKa = 4.32EE148 pKa = 4.36EE149 pKa = 4.17EE150 pKa = 4.63NEE152 pKa = 4.18VLEE155 pKa = 4.32NCVNCDD161 pKa = 3.21EE162 pKa = 5.2EE163 pKa = 5.05YY164 pKa = 11.45NDD166 pKa = 4.02VNDD169 pKa = 4.63DD170 pKa = 3.65VLIIPLCWDD179 pKa = 3.52SLQLDD184 pKa = 3.97EE185 pKa = 5.78DD186 pKa = 3.67NKK188 pKa = 10.8EE189 pKa = 4.11RR190 pKa = 11.84IEE192 pKa = 4.13DD193 pKa = 4.08FEE195 pKa = 4.4WEE197 pKa = 4.23EE198 pKa = 4.3VNVDD202 pKa = 3.14GRR204 pKa = 11.84VDD206 pKa = 3.49EE207 pKa = 5.06RR208 pKa = 11.84EE209 pKa = 4.18VLSMLTIDD217 pKa = 4.63DD218 pKa = 4.32DD219 pKa = 5.27RR220 pKa = 11.84SVSVSGIPVIEE231 pKa = 4.29EE232 pKa = 3.9EE233 pKa = 4.38EE234 pKa = 4.15EE235 pKa = 4.15DD236 pKa = 3.85VSVVRR241 pKa = 11.84MGGMEE246 pKa = 3.96NLEE249 pKa = 4.05WEE251 pKa = 4.61VLLNANNLNTITSPDD266 pKa = 4.0LDD268 pKa = 3.89NDD270 pKa = 4.15DD271 pKa = 5.48DD272 pKa = 5.04LEE274 pKa = 4.16EE275 pKa = 5.31PYY277 pKa = 10.75FSDD280 pKa = 3.1HH281 pKa = 7.71DD282 pKa = 4.18EE283 pKa = 4.53YY284 pKa = 10.84IYY286 pKa = 10.69ATDD289 pKa = 3.68YY290 pKa = 11.86EE291 pKa = 4.67MMFGQFVEE299 pKa = 4.08NEE301 pKa = 4.01YY302 pKa = 9.16TLRR305 pKa = 11.84PPASVSVVRR314 pKa = 11.84NLPSVVVTKK323 pKa = 10.93EE324 pKa = 4.2DD325 pKa = 3.26VDD327 pKa = 4.07NNNALCAVCKK337 pKa = 10.5DD338 pKa = 3.73DD339 pKa = 5.22FAVAEE344 pKa = 4.13QAKK347 pKa = 9.56QLPCSHH353 pKa = 7.38RR354 pKa = 11.84YY355 pKa = 9.56HH356 pKa = 7.26GDD358 pKa = 3.72CIVPWLAIRR367 pKa = 11.84NTCPVCRR374 pKa = 11.84YY375 pKa = 7.43EE376 pKa = 5.81FPTDD380 pKa = 3.41DD381 pKa = 3.87ADD383 pKa = 3.92YY384 pKa = 11.43EE385 pKa = 4.44RR386 pKa = 11.84MRR388 pKa = 11.84TNRR391 pKa = 11.84SALRR395 pKa = 11.84AA396 pKa = 3.67
MM1 pKa = 7.76ADD3 pKa = 2.79VTTYY7 pKa = 10.69LRR9 pKa = 11.84HH10 pKa = 6.25HH11 pKa = 6.76KK12 pKa = 9.93PDD14 pKa = 3.57EE15 pKa = 4.68DD16 pKa = 4.19FDD18 pKa = 6.28SITLPYY24 pKa = 9.97FSQSSPDD31 pKa = 3.59FNFDD35 pKa = 3.88FYY37 pKa = 11.45SSEE40 pKa = 4.31PEE42 pKa = 4.01FPPTPEE48 pKa = 4.94PDD50 pKa = 3.49SLSLFDD56 pKa = 4.57RR57 pKa = 11.84HH58 pKa = 5.39NQINFAIDD66 pKa = 3.54IFQQCVDD73 pKa = 3.39QSQVSGHH80 pKa = 6.3GYY82 pKa = 9.93FVTEE86 pKa = 4.54PRR88 pKa = 11.84NDD90 pKa = 3.74DD91 pKa = 3.63DD92 pKa = 5.43FGVIDD97 pKa = 4.58VYY99 pKa = 11.45SALDD103 pKa = 3.34MDD105 pKa = 5.88DD106 pKa = 5.14NLGLDD111 pKa = 3.45LGFGVEE117 pKa = 4.18RR118 pKa = 11.84DD119 pKa = 3.55GVSISEE125 pKa = 4.22SNEE128 pKa = 3.81APFHH132 pKa = 5.92NCVCVMGLGSDD143 pKa = 3.42SDD145 pKa = 3.89EE146 pKa = 4.1EE147 pKa = 4.32EE148 pKa = 4.36EE149 pKa = 4.17EE150 pKa = 4.63NEE152 pKa = 4.18VLEE155 pKa = 4.32NCVNCDD161 pKa = 3.21EE162 pKa = 5.2EE163 pKa = 5.05YY164 pKa = 11.45NDD166 pKa = 4.02VNDD169 pKa = 4.63DD170 pKa = 3.65VLIIPLCWDD179 pKa = 3.52SLQLDD184 pKa = 3.97EE185 pKa = 5.78DD186 pKa = 3.67NKK188 pKa = 10.8EE189 pKa = 4.11RR190 pKa = 11.84IEE192 pKa = 4.13DD193 pKa = 4.08FEE195 pKa = 4.4WEE197 pKa = 4.23EE198 pKa = 4.3VNVDD202 pKa = 3.14GRR204 pKa = 11.84VDD206 pKa = 3.49EE207 pKa = 5.06RR208 pKa = 11.84EE209 pKa = 4.18VLSMLTIDD217 pKa = 4.63DD218 pKa = 4.32DD219 pKa = 5.27RR220 pKa = 11.84SVSVSGIPVIEE231 pKa = 4.29EE232 pKa = 3.9EE233 pKa = 4.38EE234 pKa = 4.15EE235 pKa = 4.15DD236 pKa = 3.85VSVVRR241 pKa = 11.84MGGMEE246 pKa = 3.96NLEE249 pKa = 4.05WEE251 pKa = 4.61VLLNANNLNTITSPDD266 pKa = 4.0LDD268 pKa = 3.89NDD270 pKa = 4.15DD271 pKa = 5.48DD272 pKa = 5.04LEE274 pKa = 4.16EE275 pKa = 5.31PYY277 pKa = 10.75FSDD280 pKa = 3.1HH281 pKa = 7.71DD282 pKa = 4.18EE283 pKa = 4.53YY284 pKa = 10.84IYY286 pKa = 10.69ATDD289 pKa = 3.68YY290 pKa = 11.86EE291 pKa = 4.67MMFGQFVEE299 pKa = 4.08NEE301 pKa = 4.01YY302 pKa = 9.16TLRR305 pKa = 11.84PPASVSVVRR314 pKa = 11.84NLPSVVVTKK323 pKa = 10.93EE324 pKa = 4.2DD325 pKa = 3.26VDD327 pKa = 4.07NNNALCAVCKK337 pKa = 10.5DD338 pKa = 3.73DD339 pKa = 5.22FAVAEE344 pKa = 4.13QAKK347 pKa = 9.56QLPCSHH353 pKa = 7.38RR354 pKa = 11.84YY355 pKa = 9.56HH356 pKa = 7.26GDD358 pKa = 3.72CIVPWLAIRR367 pKa = 11.84NTCPVCRR374 pKa = 11.84YY375 pKa = 7.43EE376 pKa = 5.81FPTDD380 pKa = 3.41DD381 pKa = 3.87ADD383 pKa = 3.92YY384 pKa = 11.43EE385 pKa = 4.44RR386 pKa = 11.84MRR388 pKa = 11.84TNRR391 pKa = 11.84SALRR395 pKa = 11.84AA396 pKa = 3.67
Molecular weight: 45.22 kDa
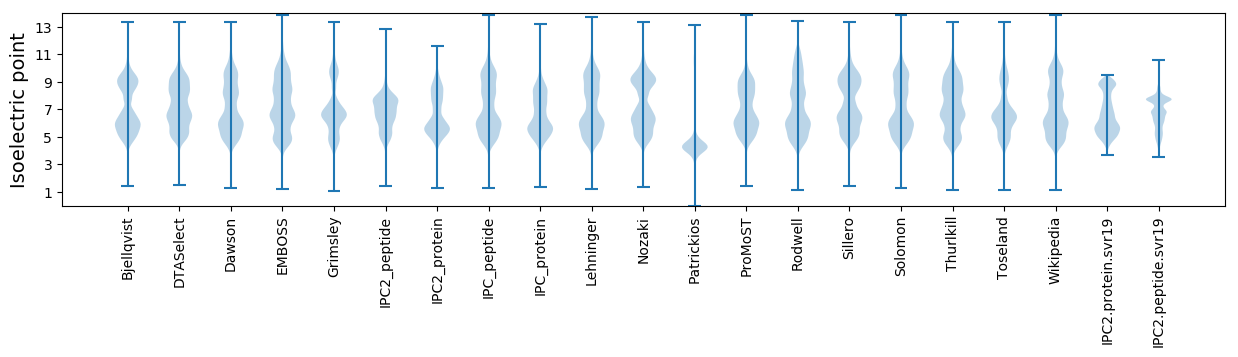
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J7HC80|A0A1J7HC80_LUPAN AB hydrolase-1 domain-containing protein OS=Lupinus angustifolius OX=3871 GN=TanjilG_24992 PE=4 SV=1
MM1 pKa = 7.09FRR3 pKa = 11.84VLEE6 pKa = 4.19FQCFDD11 pKa = 3.21GCGVEE16 pKa = 4.52GRR18 pKa = 11.84GSRR21 pKa = 11.84VQGPRR26 pKa = 11.84TRR28 pKa = 11.84VYY30 pKa = 10.53GLRR33 pKa = 11.84SRR35 pKa = 11.84VQGRR39 pKa = 11.84GSRR42 pKa = 11.84VQGPRR47 pKa = 11.84STVHH51 pKa = 6.22GPGSTVHH58 pKa = 6.94GIGSRR63 pKa = 11.84VQGPGSRR70 pKa = 11.84VQGPGSRR77 pKa = 11.84VQGPGSRR84 pKa = 11.84VQGPGSRR91 pKa = 11.84VQGPGSRR98 pKa = 11.84VQGPGSRR105 pKa = 11.84VQGPGSRR112 pKa = 11.84VQGPGSRR119 pKa = 11.84VQGPGSRR126 pKa = 11.84VQGPGSRR133 pKa = 11.84VQGPGSRR140 pKa = 11.84VQGPGSRR147 pKa = 11.84VQGPGSRR154 pKa = 11.84VQGPGSRR161 pKa = 11.84VQGPGSRR168 pKa = 11.84VQGPGSRR175 pKa = 11.84VQGPGSRR182 pKa = 11.84VQGPGSRR189 pKa = 11.84VQGPGSRR196 pKa = 11.84VQGPGSRR203 pKa = 11.84VQGPGSRR210 pKa = 11.84VQGPGSRR217 pKa = 11.84VQGPGSRR224 pKa = 11.84VQGPGSRR231 pKa = 11.84VQGPGSRR238 pKa = 11.84VQGPGSRR245 pKa = 11.84VQGPGSRR252 pKa = 11.84VQGPGSRR259 pKa = 11.84VQGPGSRR266 pKa = 11.84VQGPGSRR273 pKa = 11.84VQGPGSRR280 pKa = 11.84VQGPGSRR287 pKa = 11.84VQGPGSRR294 pKa = 11.84VQGPGSRR301 pKa = 11.84VQGPGSRR308 pKa = 11.84VQGPGSRR315 pKa = 11.84VQGPGSRR322 pKa = 11.84VQGPGSRR329 pKa = 11.84VQGPGSRR336 pKa = 11.84VQGPGSRR343 pKa = 11.84VQGPGSRR350 pKa = 11.84VQGPGSRR357 pKa = 11.84VQGPGSRR364 pKa = 11.84VQGPGSRR371 pKa = 11.84VQGPGSRR378 pKa = 11.84VQGPGSRR385 pKa = 11.84VQGPGSRR392 pKa = 11.84VQGPGSRR399 pKa = 11.84VQGPGSRR406 pKa = 11.84VQGPGSRR413 pKa = 11.84VQGPGSRR420 pKa = 11.84VQGPGSRR427 pKa = 11.84VQGPGSRR434 pKa = 11.84VQGPGSRR441 pKa = 11.84VQGPGSRR448 pKa = 11.84VQGPGSRR455 pKa = 11.84VQGPGSRR462 pKa = 11.84VQGPGSRR469 pKa = 11.84VQGPGSRR476 pKa = 11.84VQGPGSRR483 pKa = 11.84VQGPGSRR490 pKa = 11.84VQGPGSRR497 pKa = 11.84VQGPGSRR504 pKa = 11.84VQGPGSRR511 pKa = 11.84VQGPGSRR518 pKa = 11.84VQGPGSRR525 pKa = 11.84VQGPGSRR532 pKa = 11.84VQGPGSRR539 pKa = 11.84VQGPGSRR546 pKa = 11.84VQGPGSRR553 pKa = 11.84VQGPGSRR560 pKa = 11.84VQGPGSRR567 pKa = 11.84VQGPGSRR574 pKa = 11.84VQGPGSRR581 pKa = 11.84VQGPGSRR588 pKa = 11.84VQGPGSRR595 pKa = 11.84VQGPGSRR602 pKa = 11.84VQGPGSRR609 pKa = 11.84VQGPGSRR616 pKa = 11.84VQGPGSRR623 pKa = 11.84VQGPGSRR630 pKa = 11.84VQGPGSRR637 pKa = 11.84VQGPGSRR644 pKa = 11.84VQGPGSRR651 pKa = 11.84VQGPGSRR658 pKa = 11.84VQGPGSRR665 pKa = 11.84VQGPGSRR672 pKa = 11.84VQGPGSRR679 pKa = 11.84VQGPGSRR686 pKa = 11.84VQGPGSRR693 pKa = 11.84VQGPGSRR700 pKa = 11.84VQGPGSRR707 pKa = 11.84VQGPGSRR714 pKa = 11.84VQGPGSRR721 pKa = 11.84VQGPGSRR728 pKa = 11.84VQGPGSRR735 pKa = 11.84VQGPGSRR742 pKa = 11.84VQGPGSRR749 pKa = 11.84VQGPGSRR756 pKa = 11.84VQGPGSRR763 pKa = 11.84VQGPGSRR770 pKa = 11.84VQGPGSRR777 pKa = 11.84VQGPGSRR784 pKa = 11.84VQGPGSRR791 pKa = 11.84VQGPGSRR798 pKa = 11.84VQGPGSRR805 pKa = 11.84VQGPGSRR812 pKa = 11.84VQGPGSRR819 pKa = 11.84VQGPGSRR826 pKa = 11.84VQGPGSRR833 pKa = 11.84VQGPGSRR840 pKa = 11.84VQGPGSRR847 pKa = 11.84VQGPGSRR854 pKa = 11.84VQGPGSRR861 pKa = 11.84VQGPGSRR868 pKa = 11.84VQGPGSRR875 pKa = 11.84VQGPGSRR882 pKa = 11.84VQGPGSRR889 pKa = 11.84VQGPGSRR896 pKa = 11.84VQGPGSRR903 pKa = 11.84VQGPGSRR910 pKa = 11.84VQGPGSRR917 pKa = 11.84VQGPGSRR924 pKa = 11.84VQGPGSRR931 pKa = 11.84VQGPGSRR938 pKa = 11.84VQGPGSRR945 pKa = 11.84VQGPGSRR952 pKa = 11.84VQGPGSRR959 pKa = 11.84VQGPGSRR966 pKa = 11.84VQGPGSRR973 pKa = 11.84VQGPGSRR980 pKa = 11.84VQGPGSRR987 pKa = 11.84VQGPGSRR994 pKa = 11.84VQGPGSRR1001 pKa = 11.84VQGPGSRR1008 pKa = 11.84VQGPGSRR1015 pKa = 11.84VQGPGSRR1022 pKa = 11.84VQGPGSRR1029 pKa = 11.84VQGPGSRR1036 pKa = 11.84VQGPGSRR1043 pKa = 11.84VQGPGSRR1050 pKa = 11.84VQGPGSRR1057 pKa = 11.84VQGPGSRR1064 pKa = 11.84VQGPGSRR1071 pKa = 11.84VQGPGSRR1078 pKa = 11.84VQGPGSRR1085 pKa = 11.84VQGPGSRR1092 pKa = 11.84VQGPGSRR1099 pKa = 11.84VQGPGSRR1106 pKa = 11.84VQGPGSRR1113 pKa = 11.84VQGPGSRR1120 pKa = 11.84VQGPGSRR1127 pKa = 11.84VQGPGSRR1134 pKa = 11.84VQGPGSRR1141 pKa = 11.84VQGPGSRR1148 pKa = 11.84VQGPGSRR1155 pKa = 11.84VQGPGSRR1162 pKa = 11.84VQGPGSRR1169 pKa = 11.84VQGPGSRR1176 pKa = 11.84VQGPGSRR1183 pKa = 11.84VQGPGSRR1190 pKa = 11.84VQGPGSRR1197 pKa = 11.84VQGPGSRR1204 pKa = 11.84VQGPGSRR1211 pKa = 11.84VQGPGSRR1218 pKa = 11.84VQGPGSRR1225 pKa = 11.84VQGPGSRR1232 pKa = 11.84VQGPGSRR1239 pKa = 11.84VQGPGSRR1246 pKa = 11.84VQGPGSRR1253 pKa = 11.84VQGPGSRR1260 pKa = 11.84VQGPGSRR1267 pKa = 11.84VQGPGSRR1274 pKa = 11.84VQGPGSRR1281 pKa = 11.84NYY1283 pKa = 10.89
MM1 pKa = 7.09FRR3 pKa = 11.84VLEE6 pKa = 4.19FQCFDD11 pKa = 3.21GCGVEE16 pKa = 4.52GRR18 pKa = 11.84GSRR21 pKa = 11.84VQGPRR26 pKa = 11.84TRR28 pKa = 11.84VYY30 pKa = 10.53GLRR33 pKa = 11.84SRR35 pKa = 11.84VQGRR39 pKa = 11.84GSRR42 pKa = 11.84VQGPRR47 pKa = 11.84STVHH51 pKa = 6.22GPGSTVHH58 pKa = 6.94GIGSRR63 pKa = 11.84VQGPGSRR70 pKa = 11.84VQGPGSRR77 pKa = 11.84VQGPGSRR84 pKa = 11.84VQGPGSRR91 pKa = 11.84VQGPGSRR98 pKa = 11.84VQGPGSRR105 pKa = 11.84VQGPGSRR112 pKa = 11.84VQGPGSRR119 pKa = 11.84VQGPGSRR126 pKa = 11.84VQGPGSRR133 pKa = 11.84VQGPGSRR140 pKa = 11.84VQGPGSRR147 pKa = 11.84VQGPGSRR154 pKa = 11.84VQGPGSRR161 pKa = 11.84VQGPGSRR168 pKa = 11.84VQGPGSRR175 pKa = 11.84VQGPGSRR182 pKa = 11.84VQGPGSRR189 pKa = 11.84VQGPGSRR196 pKa = 11.84VQGPGSRR203 pKa = 11.84VQGPGSRR210 pKa = 11.84VQGPGSRR217 pKa = 11.84VQGPGSRR224 pKa = 11.84VQGPGSRR231 pKa = 11.84VQGPGSRR238 pKa = 11.84VQGPGSRR245 pKa = 11.84VQGPGSRR252 pKa = 11.84VQGPGSRR259 pKa = 11.84VQGPGSRR266 pKa = 11.84VQGPGSRR273 pKa = 11.84VQGPGSRR280 pKa = 11.84VQGPGSRR287 pKa = 11.84VQGPGSRR294 pKa = 11.84VQGPGSRR301 pKa = 11.84VQGPGSRR308 pKa = 11.84VQGPGSRR315 pKa = 11.84VQGPGSRR322 pKa = 11.84VQGPGSRR329 pKa = 11.84VQGPGSRR336 pKa = 11.84VQGPGSRR343 pKa = 11.84VQGPGSRR350 pKa = 11.84VQGPGSRR357 pKa = 11.84VQGPGSRR364 pKa = 11.84VQGPGSRR371 pKa = 11.84VQGPGSRR378 pKa = 11.84VQGPGSRR385 pKa = 11.84VQGPGSRR392 pKa = 11.84VQGPGSRR399 pKa = 11.84VQGPGSRR406 pKa = 11.84VQGPGSRR413 pKa = 11.84VQGPGSRR420 pKa = 11.84VQGPGSRR427 pKa = 11.84VQGPGSRR434 pKa = 11.84VQGPGSRR441 pKa = 11.84VQGPGSRR448 pKa = 11.84VQGPGSRR455 pKa = 11.84VQGPGSRR462 pKa = 11.84VQGPGSRR469 pKa = 11.84VQGPGSRR476 pKa = 11.84VQGPGSRR483 pKa = 11.84VQGPGSRR490 pKa = 11.84VQGPGSRR497 pKa = 11.84VQGPGSRR504 pKa = 11.84VQGPGSRR511 pKa = 11.84VQGPGSRR518 pKa = 11.84VQGPGSRR525 pKa = 11.84VQGPGSRR532 pKa = 11.84VQGPGSRR539 pKa = 11.84VQGPGSRR546 pKa = 11.84VQGPGSRR553 pKa = 11.84VQGPGSRR560 pKa = 11.84VQGPGSRR567 pKa = 11.84VQGPGSRR574 pKa = 11.84VQGPGSRR581 pKa = 11.84VQGPGSRR588 pKa = 11.84VQGPGSRR595 pKa = 11.84VQGPGSRR602 pKa = 11.84VQGPGSRR609 pKa = 11.84VQGPGSRR616 pKa = 11.84VQGPGSRR623 pKa = 11.84VQGPGSRR630 pKa = 11.84VQGPGSRR637 pKa = 11.84VQGPGSRR644 pKa = 11.84VQGPGSRR651 pKa = 11.84VQGPGSRR658 pKa = 11.84VQGPGSRR665 pKa = 11.84VQGPGSRR672 pKa = 11.84VQGPGSRR679 pKa = 11.84VQGPGSRR686 pKa = 11.84VQGPGSRR693 pKa = 11.84VQGPGSRR700 pKa = 11.84VQGPGSRR707 pKa = 11.84VQGPGSRR714 pKa = 11.84VQGPGSRR721 pKa = 11.84VQGPGSRR728 pKa = 11.84VQGPGSRR735 pKa = 11.84VQGPGSRR742 pKa = 11.84VQGPGSRR749 pKa = 11.84VQGPGSRR756 pKa = 11.84VQGPGSRR763 pKa = 11.84VQGPGSRR770 pKa = 11.84VQGPGSRR777 pKa = 11.84VQGPGSRR784 pKa = 11.84VQGPGSRR791 pKa = 11.84VQGPGSRR798 pKa = 11.84VQGPGSRR805 pKa = 11.84VQGPGSRR812 pKa = 11.84VQGPGSRR819 pKa = 11.84VQGPGSRR826 pKa = 11.84VQGPGSRR833 pKa = 11.84VQGPGSRR840 pKa = 11.84VQGPGSRR847 pKa = 11.84VQGPGSRR854 pKa = 11.84VQGPGSRR861 pKa = 11.84VQGPGSRR868 pKa = 11.84VQGPGSRR875 pKa = 11.84VQGPGSRR882 pKa = 11.84VQGPGSRR889 pKa = 11.84VQGPGSRR896 pKa = 11.84VQGPGSRR903 pKa = 11.84VQGPGSRR910 pKa = 11.84VQGPGSRR917 pKa = 11.84VQGPGSRR924 pKa = 11.84VQGPGSRR931 pKa = 11.84VQGPGSRR938 pKa = 11.84VQGPGSRR945 pKa = 11.84VQGPGSRR952 pKa = 11.84VQGPGSRR959 pKa = 11.84VQGPGSRR966 pKa = 11.84VQGPGSRR973 pKa = 11.84VQGPGSRR980 pKa = 11.84VQGPGSRR987 pKa = 11.84VQGPGSRR994 pKa = 11.84VQGPGSRR1001 pKa = 11.84VQGPGSRR1008 pKa = 11.84VQGPGSRR1015 pKa = 11.84VQGPGSRR1022 pKa = 11.84VQGPGSRR1029 pKa = 11.84VQGPGSRR1036 pKa = 11.84VQGPGSRR1043 pKa = 11.84VQGPGSRR1050 pKa = 11.84VQGPGSRR1057 pKa = 11.84VQGPGSRR1064 pKa = 11.84VQGPGSRR1071 pKa = 11.84VQGPGSRR1078 pKa = 11.84VQGPGSRR1085 pKa = 11.84VQGPGSRR1092 pKa = 11.84VQGPGSRR1099 pKa = 11.84VQGPGSRR1106 pKa = 11.84VQGPGSRR1113 pKa = 11.84VQGPGSRR1120 pKa = 11.84VQGPGSRR1127 pKa = 11.84VQGPGSRR1134 pKa = 11.84VQGPGSRR1141 pKa = 11.84VQGPGSRR1148 pKa = 11.84VQGPGSRR1155 pKa = 11.84VQGPGSRR1162 pKa = 11.84VQGPGSRR1169 pKa = 11.84VQGPGSRR1176 pKa = 11.84VQGPGSRR1183 pKa = 11.84VQGPGSRR1190 pKa = 11.84VQGPGSRR1197 pKa = 11.84VQGPGSRR1204 pKa = 11.84VQGPGSRR1211 pKa = 11.84VQGPGSRR1218 pKa = 11.84VQGPGSRR1225 pKa = 11.84VQGPGSRR1232 pKa = 11.84VQGPGSRR1239 pKa = 11.84VQGPGSRR1246 pKa = 11.84VQGPGSRR1253 pKa = 11.84VQGPGSRR1260 pKa = 11.84VQGPGSRR1267 pKa = 11.84VQGPGSRR1274 pKa = 11.84VQGPGSRR1281 pKa = 11.84NYY1283 pKa = 10.89
Molecular weight: 125.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13402789 |
49 |
5685 |
430.9 |
48.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.449 ± 0.013 | 1.745 ± 0.007 |
5.389 ± 0.012 | 6.518 ± 0.022 |
4.151 ± 0.01 | 6.434 ± 0.015 |
2.534 ± 0.006 | 5.463 ± 0.01 |
6.276 ± 0.015 | 9.382 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.393 ± 0.006 | 4.903 ± 0.013 |
4.929 ± 0.02 | 3.728 ± 0.01 |
4.975 ± 0.011 | 9.129 ± 0.018 |
5.011 ± 0.008 | 6.543 ± 0.01 |
1.21 ± 0.005 | 2.838 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
