
Changjiang narna-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
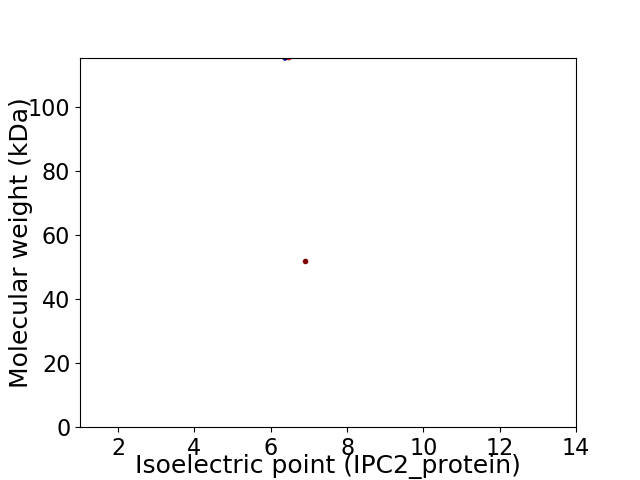
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIH4|A0A1L3KIH4_9VIRU Capsid protein OS=Changjiang narna-like virus 2 OX=1922777 PE=3 SV=1
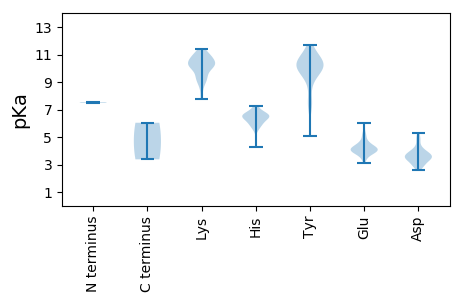
MM1 pKa = 7.49NSASCTNYY9 pKa = 9.91TVIHH13 pKa = 7.14LIDD16 pKa = 5.13GILIRR21 pKa = 11.84LHH23 pKa = 6.06AVRR26 pKa = 11.84PEE28 pKa = 3.19WKK30 pKa = 9.24YY31 pKa = 9.72RR32 pKa = 11.84HH33 pKa = 5.95GGPRR37 pKa = 11.84QWLYY41 pKa = 10.21VAEE44 pKa = 4.71PKK46 pKa = 10.3TSFEE50 pKa = 4.15VEE52 pKa = 4.03LAQVATTLSDD62 pKa = 3.47PSFPPSEE69 pKa = 4.04EE70 pKa = 3.8RR71 pKa = 11.84VKK73 pKa = 11.17GGGYY77 pKa = 10.01RR78 pKa = 11.84LPDD81 pKa = 3.39GTVTMVPPPPKK92 pKa = 10.13LEE94 pKa = 4.28SGAALPAPKK103 pKa = 9.42PGQVHH108 pKa = 6.7HH109 pKa = 6.49SVDD112 pKa = 3.02WRR114 pKa = 11.84LLHH117 pKa = 6.49ILSGLRR123 pKa = 11.84AVLLVLVDD131 pKa = 3.84SCGSCAQSPDD141 pKa = 3.53FVGNQPWTDD150 pKa = 3.76SIAVIKK156 pKa = 10.67AMRR159 pKa = 11.84SWSEE163 pKa = 3.5EE164 pKa = 3.8DD165 pKa = 3.5FVPNAKK171 pKa = 9.25YY172 pKa = 9.98WKK174 pKa = 8.97DD175 pKa = 3.04WPLARR180 pKa = 11.84LLKK183 pKa = 10.1QQLPNRR189 pKa = 11.84PPSWKK194 pKa = 9.35YY195 pKa = 11.22GPMACLFTGEE205 pKa = 4.25TGRR208 pKa = 11.84YY209 pKa = 5.1WNRR212 pKa = 11.84IATYY216 pKa = 8.21DD217 pKa = 3.37TTRR220 pKa = 11.84PDD222 pKa = 2.88SFVFYY227 pKa = 10.3RR228 pKa = 11.84AAFGLSQSKK237 pKa = 10.45RR238 pKa = 11.84GFAAVPRR245 pKa = 11.84SFVKK249 pKa = 10.68RR250 pKa = 11.84SLEE253 pKa = 3.6KK254 pKa = 10.2HH255 pKa = 5.71ARR257 pKa = 11.84QLSTPPTSDD266 pKa = 3.58PDD268 pKa = 3.62MEE270 pKa = 4.43SARR273 pKa = 11.84IFAEE277 pKa = 4.11TFFEE281 pKa = 4.7GFRR284 pKa = 11.84IPDD287 pKa = 3.59ILKK290 pKa = 10.15SLPDD294 pKa = 3.23IEE296 pKa = 4.64GTLRR300 pKa = 11.84ACVEE304 pKa = 4.27NPRR307 pKa = 11.84HH308 pKa = 5.81AGGARR313 pKa = 11.84EE314 pKa = 4.26FLRR317 pKa = 11.84HH318 pKa = 6.39LARR321 pKa = 11.84KK322 pKa = 7.74HH323 pKa = 6.13HH324 pKa = 6.19GMLDD328 pKa = 4.03SYY330 pKa = 10.09PVSYY334 pKa = 10.75SGLASVLEE342 pKa = 4.4EE343 pKa = 4.22TPEE346 pKa = 4.0ALPVYY351 pKa = 10.24VKK353 pKa = 9.97TRR355 pKa = 11.84EE356 pKa = 3.88PLVRR360 pKa = 11.84MIEE363 pKa = 3.85ISPGVIVEE371 pKa = 4.06EE372 pKa = 4.73HH373 pKa = 6.51GLPPLSPEE381 pKa = 3.81DD382 pKa = 3.23WRR384 pKa = 11.84VLVNNHH390 pKa = 5.5SPQNARR396 pKa = 11.84EE397 pKa = 3.91YY398 pKa = 10.65LPPGVQDD405 pKa = 3.57AVDD408 pKa = 3.73RR409 pKa = 11.84LIDD412 pKa = 3.89SEE414 pKa = 4.53PEE416 pKa = 3.57ATNFPTCKK424 pKa = 9.5VAEE427 pKa = 4.19VLEE430 pKa = 4.12PLKK433 pKa = 11.07VRR435 pKa = 11.84LITAMDD441 pKa = 4.33ALRR444 pKa = 11.84THH446 pKa = 6.48VARR449 pKa = 11.84PLQAALWRR457 pKa = 11.84YY458 pKa = 9.39LRR460 pKa = 11.84ASPVFALIGEE470 pKa = 5.01PISEE474 pKa = 5.31DD475 pKa = 3.76LLQDD479 pKa = 3.58LCARR483 pKa = 11.84HH484 pKa = 6.04LADD487 pKa = 5.02GGGSNDD493 pKa = 2.88PWASGDD499 pKa = 3.57YY500 pKa = 10.61SAATDD505 pKa = 3.71GLDD508 pKa = 3.09IRR510 pKa = 11.84FSRR513 pKa = 11.84LVIEE517 pKa = 4.52VVLSKK522 pKa = 10.93LRR524 pKa = 11.84TEE526 pKa = 4.98DD527 pKa = 2.75KK528 pKa = 10.34WLAPFLASILDD539 pKa = 3.82QQIITYY545 pKa = 7.31PAWAKK550 pKa = 10.72LPAILQKK557 pKa = 10.7NGQLMGSVLSFPVLCLANFFAFIQSRR583 pKa = 11.84PNAAAILRR591 pKa = 11.84SRR593 pKa = 11.84RR594 pKa = 11.84LMDD597 pKa = 3.84RR598 pKa = 11.84QPVLINGDD606 pKa = 4.26DD607 pKa = 3.35ILFRR611 pKa = 11.84ATEE614 pKa = 3.79EE615 pKa = 4.34RR616 pKa = 11.84YY617 pKa = 9.78KK618 pKa = 11.06VWLSEE623 pKa = 3.74IAKK626 pKa = 10.08VGFVPSVGKK635 pKa = 10.26NFFHH639 pKa = 7.28PRR641 pKa = 11.84FFTVNSVPIEE651 pKa = 4.07YY652 pKa = 10.23LPSPTPYY659 pKa = 10.4QFWSQFSWADD669 pKa = 3.47MEE671 pKa = 5.33EE672 pKa = 4.83IAAPWNINQAPRR684 pKa = 11.84ISIRR688 pKa = 11.84GFLNVGLLTGQAKK701 pKa = 8.73LTGRR705 pKa = 11.84EE706 pKa = 4.0ALGALPLAGWHH717 pKa = 6.38AGSVLEE723 pKa = 5.24ALNPLQAHH731 pKa = 6.53KK732 pKa = 10.41WFLHH736 pKa = 4.24YY737 pKa = 10.31HH738 pKa = 5.73LEE740 pKa = 4.45EE741 pKa = 4.03IQRR744 pKa = 11.84QTRR747 pKa = 11.84FGSTTLNIFAHH758 pKa = 6.58PLLGGLGFQIPVGVEE773 pKa = 3.6PRR775 pKa = 11.84FSPEE779 pKa = 3.14QRR781 pKa = 11.84RR782 pKa = 11.84IARR785 pKa = 11.84ALFLSASASYY795 pKa = 10.1EE796 pKa = 3.93GQEE799 pKa = 4.64SRR801 pKa = 11.84FEE803 pKa = 4.3LDD805 pKa = 3.02SLLFLEE811 pKa = 5.08SRR813 pKa = 11.84TAAPLSSLGNRR824 pKa = 11.84RR825 pKa = 11.84RR826 pKa = 11.84RR827 pKa = 11.84VEE829 pKa = 3.89VEE831 pKa = 4.25LYY833 pKa = 7.73PTGTPLPEE841 pKa = 5.15GYY843 pKa = 10.44SPFTDD848 pKa = 3.32STGVQPLAMVHH859 pKa = 6.57DD860 pKa = 5.28LPSSEE865 pKa = 5.77DD866 pKa = 3.84DD867 pKa = 3.85LPTIARR873 pKa = 11.84CRR875 pKa = 11.84LSSRR879 pKa = 11.84RR880 pKa = 11.84IRR882 pKa = 11.84QLIHH886 pKa = 6.93RR887 pKa = 11.84FDD889 pKa = 4.09DD890 pKa = 4.1TVDD893 pKa = 3.63LHH895 pKa = 6.58PLEE898 pKa = 5.8LMTQFPYY905 pKa = 10.49TPVRR909 pKa = 11.84VTRR912 pKa = 11.84SEE914 pKa = 4.43LIADD918 pKa = 3.78KK919 pKa = 10.96GVVIPEE925 pKa = 3.84EE926 pKa = 4.24RR927 pKa = 11.84PFSKK931 pKa = 10.24VYY933 pKa = 10.86SMEE936 pKa = 4.22IPFQDD941 pKa = 3.5VPVPEE946 pKa = 4.46AAEE949 pKa = 4.16EE950 pKa = 3.99PVLPKK955 pKa = 10.49VYY957 pKa = 10.23EE958 pKa = 4.23PEE960 pKa = 3.89DD961 pKa = 3.52WEE963 pKa = 4.38TATVSLRR970 pKa = 11.84VINPIVAPYY979 pKa = 9.93VPPPLSPEE987 pKa = 3.34EE988 pKa = 3.92RR989 pKa = 11.84YY990 pKa = 10.21RR991 pKa = 11.84RR992 pKa = 11.84MEE994 pKa = 3.5GRR996 pKa = 11.84RR997 pKa = 11.84RR998 pKa = 11.84QVLSRR1003 pKa = 11.84EE1004 pKa = 3.8QNARR1008 pKa = 11.84NRR1010 pKa = 11.84GMIYY1014 pKa = 10.29ARR1016 pKa = 11.84SALEE1020 pKa = 3.42QSYY1023 pKa = 10.36FEE1025 pKa = 4.23II1026 pKa = 6.04
MM1 pKa = 7.49NSASCTNYY9 pKa = 9.91TVIHH13 pKa = 7.14LIDD16 pKa = 5.13GILIRR21 pKa = 11.84LHH23 pKa = 6.06AVRR26 pKa = 11.84PEE28 pKa = 3.19WKK30 pKa = 9.24YY31 pKa = 9.72RR32 pKa = 11.84HH33 pKa = 5.95GGPRR37 pKa = 11.84QWLYY41 pKa = 10.21VAEE44 pKa = 4.71PKK46 pKa = 10.3TSFEE50 pKa = 4.15VEE52 pKa = 4.03LAQVATTLSDD62 pKa = 3.47PSFPPSEE69 pKa = 4.04EE70 pKa = 3.8RR71 pKa = 11.84VKK73 pKa = 11.17GGGYY77 pKa = 10.01RR78 pKa = 11.84LPDD81 pKa = 3.39GTVTMVPPPPKK92 pKa = 10.13LEE94 pKa = 4.28SGAALPAPKK103 pKa = 9.42PGQVHH108 pKa = 6.7HH109 pKa = 6.49SVDD112 pKa = 3.02WRR114 pKa = 11.84LLHH117 pKa = 6.49ILSGLRR123 pKa = 11.84AVLLVLVDD131 pKa = 3.84SCGSCAQSPDD141 pKa = 3.53FVGNQPWTDD150 pKa = 3.76SIAVIKK156 pKa = 10.67AMRR159 pKa = 11.84SWSEE163 pKa = 3.5EE164 pKa = 3.8DD165 pKa = 3.5FVPNAKK171 pKa = 9.25YY172 pKa = 9.98WKK174 pKa = 8.97DD175 pKa = 3.04WPLARR180 pKa = 11.84LLKK183 pKa = 10.1QQLPNRR189 pKa = 11.84PPSWKK194 pKa = 9.35YY195 pKa = 11.22GPMACLFTGEE205 pKa = 4.25TGRR208 pKa = 11.84YY209 pKa = 5.1WNRR212 pKa = 11.84IATYY216 pKa = 8.21DD217 pKa = 3.37TTRR220 pKa = 11.84PDD222 pKa = 2.88SFVFYY227 pKa = 10.3RR228 pKa = 11.84AAFGLSQSKK237 pKa = 10.45RR238 pKa = 11.84GFAAVPRR245 pKa = 11.84SFVKK249 pKa = 10.68RR250 pKa = 11.84SLEE253 pKa = 3.6KK254 pKa = 10.2HH255 pKa = 5.71ARR257 pKa = 11.84QLSTPPTSDD266 pKa = 3.58PDD268 pKa = 3.62MEE270 pKa = 4.43SARR273 pKa = 11.84IFAEE277 pKa = 4.11TFFEE281 pKa = 4.7GFRR284 pKa = 11.84IPDD287 pKa = 3.59ILKK290 pKa = 10.15SLPDD294 pKa = 3.23IEE296 pKa = 4.64GTLRR300 pKa = 11.84ACVEE304 pKa = 4.27NPRR307 pKa = 11.84HH308 pKa = 5.81AGGARR313 pKa = 11.84EE314 pKa = 4.26FLRR317 pKa = 11.84HH318 pKa = 6.39LARR321 pKa = 11.84KK322 pKa = 7.74HH323 pKa = 6.13HH324 pKa = 6.19GMLDD328 pKa = 4.03SYY330 pKa = 10.09PVSYY334 pKa = 10.75SGLASVLEE342 pKa = 4.4EE343 pKa = 4.22TPEE346 pKa = 4.0ALPVYY351 pKa = 10.24VKK353 pKa = 9.97TRR355 pKa = 11.84EE356 pKa = 3.88PLVRR360 pKa = 11.84MIEE363 pKa = 3.85ISPGVIVEE371 pKa = 4.06EE372 pKa = 4.73HH373 pKa = 6.51GLPPLSPEE381 pKa = 3.81DD382 pKa = 3.23WRR384 pKa = 11.84VLVNNHH390 pKa = 5.5SPQNARR396 pKa = 11.84EE397 pKa = 3.91YY398 pKa = 10.65LPPGVQDD405 pKa = 3.57AVDD408 pKa = 3.73RR409 pKa = 11.84LIDD412 pKa = 3.89SEE414 pKa = 4.53PEE416 pKa = 3.57ATNFPTCKK424 pKa = 9.5VAEE427 pKa = 4.19VLEE430 pKa = 4.12PLKK433 pKa = 11.07VRR435 pKa = 11.84LITAMDD441 pKa = 4.33ALRR444 pKa = 11.84THH446 pKa = 6.48VARR449 pKa = 11.84PLQAALWRR457 pKa = 11.84YY458 pKa = 9.39LRR460 pKa = 11.84ASPVFALIGEE470 pKa = 5.01PISEE474 pKa = 5.31DD475 pKa = 3.76LLQDD479 pKa = 3.58LCARR483 pKa = 11.84HH484 pKa = 6.04LADD487 pKa = 5.02GGGSNDD493 pKa = 2.88PWASGDD499 pKa = 3.57YY500 pKa = 10.61SAATDD505 pKa = 3.71GLDD508 pKa = 3.09IRR510 pKa = 11.84FSRR513 pKa = 11.84LVIEE517 pKa = 4.52VVLSKK522 pKa = 10.93LRR524 pKa = 11.84TEE526 pKa = 4.98DD527 pKa = 2.75KK528 pKa = 10.34WLAPFLASILDD539 pKa = 3.82QQIITYY545 pKa = 7.31PAWAKK550 pKa = 10.72LPAILQKK557 pKa = 10.7NGQLMGSVLSFPVLCLANFFAFIQSRR583 pKa = 11.84PNAAAILRR591 pKa = 11.84SRR593 pKa = 11.84RR594 pKa = 11.84LMDD597 pKa = 3.84RR598 pKa = 11.84QPVLINGDD606 pKa = 4.26DD607 pKa = 3.35ILFRR611 pKa = 11.84ATEE614 pKa = 3.79EE615 pKa = 4.34RR616 pKa = 11.84YY617 pKa = 9.78KK618 pKa = 11.06VWLSEE623 pKa = 3.74IAKK626 pKa = 10.08VGFVPSVGKK635 pKa = 10.26NFFHH639 pKa = 7.28PRR641 pKa = 11.84FFTVNSVPIEE651 pKa = 4.07YY652 pKa = 10.23LPSPTPYY659 pKa = 10.4QFWSQFSWADD669 pKa = 3.47MEE671 pKa = 5.33EE672 pKa = 4.83IAAPWNINQAPRR684 pKa = 11.84ISIRR688 pKa = 11.84GFLNVGLLTGQAKK701 pKa = 8.73LTGRR705 pKa = 11.84EE706 pKa = 4.0ALGALPLAGWHH717 pKa = 6.38AGSVLEE723 pKa = 5.24ALNPLQAHH731 pKa = 6.53KK732 pKa = 10.41WFLHH736 pKa = 4.24YY737 pKa = 10.31HH738 pKa = 5.73LEE740 pKa = 4.45EE741 pKa = 4.03IQRR744 pKa = 11.84QTRR747 pKa = 11.84FGSTTLNIFAHH758 pKa = 6.58PLLGGLGFQIPVGVEE773 pKa = 3.6PRR775 pKa = 11.84FSPEE779 pKa = 3.14QRR781 pKa = 11.84RR782 pKa = 11.84IARR785 pKa = 11.84ALFLSASASYY795 pKa = 10.1EE796 pKa = 3.93GQEE799 pKa = 4.64SRR801 pKa = 11.84FEE803 pKa = 4.3LDD805 pKa = 3.02SLLFLEE811 pKa = 5.08SRR813 pKa = 11.84TAAPLSSLGNRR824 pKa = 11.84RR825 pKa = 11.84RR826 pKa = 11.84RR827 pKa = 11.84VEE829 pKa = 3.89VEE831 pKa = 4.25LYY833 pKa = 7.73PTGTPLPEE841 pKa = 5.15GYY843 pKa = 10.44SPFTDD848 pKa = 3.32STGVQPLAMVHH859 pKa = 6.57DD860 pKa = 5.28LPSSEE865 pKa = 5.77DD866 pKa = 3.84DD867 pKa = 3.85LPTIARR873 pKa = 11.84CRR875 pKa = 11.84LSSRR879 pKa = 11.84RR880 pKa = 11.84IRR882 pKa = 11.84QLIHH886 pKa = 6.93RR887 pKa = 11.84FDD889 pKa = 4.09DD890 pKa = 4.1TVDD893 pKa = 3.63LHH895 pKa = 6.58PLEE898 pKa = 5.8LMTQFPYY905 pKa = 10.49TPVRR909 pKa = 11.84VTRR912 pKa = 11.84SEE914 pKa = 4.43LIADD918 pKa = 3.78KK919 pKa = 10.96GVVIPEE925 pKa = 3.84EE926 pKa = 4.24RR927 pKa = 11.84PFSKK931 pKa = 10.24VYY933 pKa = 10.86SMEE936 pKa = 4.22IPFQDD941 pKa = 3.5VPVPEE946 pKa = 4.46AAEE949 pKa = 4.16EE950 pKa = 3.99PVLPKK955 pKa = 10.49VYY957 pKa = 10.23EE958 pKa = 4.23PEE960 pKa = 3.89DD961 pKa = 3.52WEE963 pKa = 4.38TATVSLRR970 pKa = 11.84VINPIVAPYY979 pKa = 9.93VPPPLSPEE987 pKa = 3.34EE988 pKa = 3.92RR989 pKa = 11.84YY990 pKa = 10.21RR991 pKa = 11.84RR992 pKa = 11.84MEE994 pKa = 3.5GRR996 pKa = 11.84RR997 pKa = 11.84RR998 pKa = 11.84QVLSRR1003 pKa = 11.84EE1004 pKa = 3.8QNARR1008 pKa = 11.84NRR1010 pKa = 11.84GMIYY1014 pKa = 10.29ARR1016 pKa = 11.84SALEE1020 pKa = 3.42QSYY1023 pKa = 10.36FEE1025 pKa = 4.23II1026 pKa = 6.04
Molecular weight: 115.47 kDa
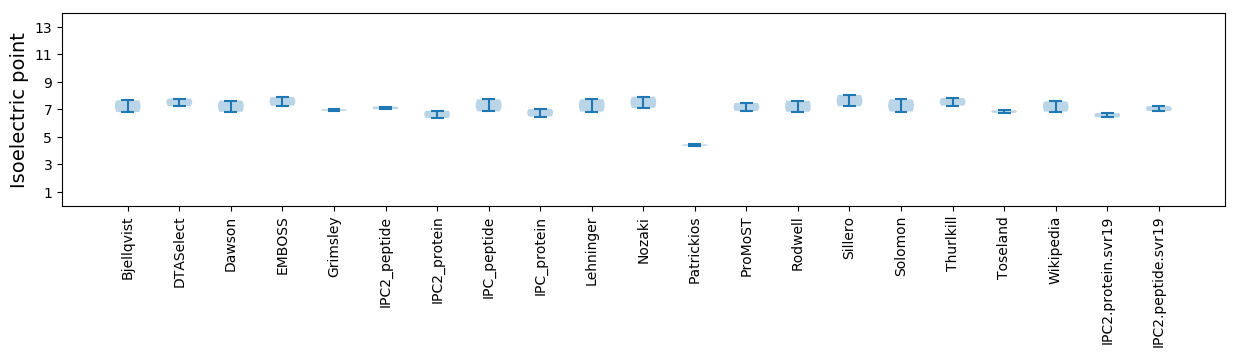
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIH4|A0A1L3KIH4_9VIRU Capsid protein OS=Changjiang narna-like virus 2 OX=1922777 PE=3 SV=1
MM1 pKa = 7.54PRR3 pKa = 11.84DD4 pKa = 3.65CTALSDD10 pKa = 3.79HH11 pKa = 6.63SALSDD16 pKa = 3.7VQSVPSGWIQYY27 pKa = 7.09TDD29 pKa = 3.33KK30 pKa = 11.43PKK32 pKa = 8.14MTKK35 pKa = 10.2RR36 pKa = 11.84NASSSRR42 pKa = 11.84ARR44 pKa = 11.84SANNRR49 pKa = 11.84TRR51 pKa = 11.84RR52 pKa = 11.84LRR54 pKa = 11.84GKK56 pKa = 10.38GDD58 pKa = 3.35YY59 pKa = 11.02SEE61 pKa = 6.01DD62 pKa = 3.01IRR64 pKa = 11.84NEE66 pKa = 4.02TNVATRR72 pKa = 11.84LEE74 pKa = 4.15KK75 pKa = 10.74KK76 pKa = 9.5IDD78 pKa = 3.36HH79 pKa = 6.75LEE81 pKa = 4.02RR82 pKa = 11.84SLVHH86 pKa = 6.05ATPSIAKK93 pKa = 8.83GASIAGRR100 pKa = 11.84ALGSLLGQGDD110 pKa = 4.82LGAMAGEE117 pKa = 4.53GLSKK121 pKa = 10.99LFGFGDD127 pKa = 3.4YY128 pKa = 10.16RR129 pKa = 11.84VAVKK133 pKa = 10.6GNSLMAGTTSTPVPKK148 pKa = 10.58FSGDD152 pKa = 2.98GRR154 pKa = 11.84RR155 pKa = 11.84GTRR158 pKa = 11.84ITEE161 pKa = 3.82RR162 pKa = 11.84EE163 pKa = 4.03FIGNVLTGSVVSGSSVFNNAVYY185 pKa = 9.53PVNPIDD191 pKa = 4.76SNTFPWLSRR200 pKa = 11.84IAQQFDD206 pKa = 2.58QWEE209 pKa = 3.84PHH211 pKa = 6.41GIVFEE216 pKa = 4.13YY217 pKa = 9.2RR218 pKa = 11.84TTSSTFNGTSQALGAVIMATDD239 pKa = 4.14YY240 pKa = 11.69DD241 pKa = 4.04SLDD244 pKa = 3.04AAFRR248 pKa = 11.84DD249 pKa = 3.81KK250 pKa = 11.18QQMEE254 pKa = 4.24NADD257 pKa = 4.09YY258 pKa = 11.14ACSTVPSQSLLHH270 pKa = 6.58GLEE273 pKa = 4.17CDD275 pKa = 3.3PRR277 pKa = 11.84EE278 pKa = 4.11RR279 pKa = 11.84PLEE282 pKa = 3.91VMYY285 pKa = 9.39TQTRR289 pKa = 11.84AGNQNFSSLGNFQIATQGCSTAGTTLGEE317 pKa = 3.9LWISYY322 pKa = 10.48DD323 pKa = 2.69ITFYY327 pKa = 11.32KK328 pKa = 10.28KK329 pKa = 10.19VLPEE333 pKa = 4.11HH334 pKa = 6.74SEE336 pKa = 3.78SSPFVNINNKK346 pKa = 9.97AIALSQLFKK355 pKa = 10.81PNSATIAKK363 pKa = 9.39DD364 pKa = 2.85ITVALIPGYY373 pKa = 7.34GTRR376 pKa = 11.84FVFPPSQSSGRR387 pKa = 11.84YY388 pKa = 9.02LFLAYY393 pKa = 9.68MSEE396 pKa = 4.22FLSGDD401 pKa = 3.45LGQIVPFGHH410 pKa = 6.67KK411 pKa = 9.96NCEE414 pKa = 4.08QVSGYY419 pKa = 11.09AGTNTGIGTPGVSSSVINITGPGAWFHH446 pKa = 6.46TGLKK450 pKa = 10.08QGTPPQEE457 pKa = 4.55ANAKK461 pKa = 9.11MILIPIDD468 pKa = 4.79FDD470 pKa = 4.52TNWDD474 pKa = 3.44VTPPVV479 pKa = 3.38
MM1 pKa = 7.54PRR3 pKa = 11.84DD4 pKa = 3.65CTALSDD10 pKa = 3.79HH11 pKa = 6.63SALSDD16 pKa = 3.7VQSVPSGWIQYY27 pKa = 7.09TDD29 pKa = 3.33KK30 pKa = 11.43PKK32 pKa = 8.14MTKK35 pKa = 10.2RR36 pKa = 11.84NASSSRR42 pKa = 11.84ARR44 pKa = 11.84SANNRR49 pKa = 11.84TRR51 pKa = 11.84RR52 pKa = 11.84LRR54 pKa = 11.84GKK56 pKa = 10.38GDD58 pKa = 3.35YY59 pKa = 11.02SEE61 pKa = 6.01DD62 pKa = 3.01IRR64 pKa = 11.84NEE66 pKa = 4.02TNVATRR72 pKa = 11.84LEE74 pKa = 4.15KK75 pKa = 10.74KK76 pKa = 9.5IDD78 pKa = 3.36HH79 pKa = 6.75LEE81 pKa = 4.02RR82 pKa = 11.84SLVHH86 pKa = 6.05ATPSIAKK93 pKa = 8.83GASIAGRR100 pKa = 11.84ALGSLLGQGDD110 pKa = 4.82LGAMAGEE117 pKa = 4.53GLSKK121 pKa = 10.99LFGFGDD127 pKa = 3.4YY128 pKa = 10.16RR129 pKa = 11.84VAVKK133 pKa = 10.6GNSLMAGTTSTPVPKK148 pKa = 10.58FSGDD152 pKa = 2.98GRR154 pKa = 11.84RR155 pKa = 11.84GTRR158 pKa = 11.84ITEE161 pKa = 3.82RR162 pKa = 11.84EE163 pKa = 4.03FIGNVLTGSVVSGSSVFNNAVYY185 pKa = 9.53PVNPIDD191 pKa = 4.76SNTFPWLSRR200 pKa = 11.84IAQQFDD206 pKa = 2.58QWEE209 pKa = 3.84PHH211 pKa = 6.41GIVFEE216 pKa = 4.13YY217 pKa = 9.2RR218 pKa = 11.84TTSSTFNGTSQALGAVIMATDD239 pKa = 4.14YY240 pKa = 11.69DD241 pKa = 4.04SLDD244 pKa = 3.04AAFRR248 pKa = 11.84DD249 pKa = 3.81KK250 pKa = 11.18QQMEE254 pKa = 4.24NADD257 pKa = 4.09YY258 pKa = 11.14ACSTVPSQSLLHH270 pKa = 6.58GLEE273 pKa = 4.17CDD275 pKa = 3.3PRR277 pKa = 11.84EE278 pKa = 4.11RR279 pKa = 11.84PLEE282 pKa = 3.91VMYY285 pKa = 9.39TQTRR289 pKa = 11.84AGNQNFSSLGNFQIATQGCSTAGTTLGEE317 pKa = 3.9LWISYY322 pKa = 10.48DD323 pKa = 2.69ITFYY327 pKa = 11.32KK328 pKa = 10.28KK329 pKa = 10.19VLPEE333 pKa = 4.11HH334 pKa = 6.74SEE336 pKa = 3.78SSPFVNINNKK346 pKa = 9.97AIALSQLFKK355 pKa = 10.81PNSATIAKK363 pKa = 9.39DD364 pKa = 2.85ITVALIPGYY373 pKa = 7.34GTRR376 pKa = 11.84FVFPPSQSSGRR387 pKa = 11.84YY388 pKa = 9.02LFLAYY393 pKa = 9.68MSEE396 pKa = 4.22FLSGDD401 pKa = 3.45LGQIVPFGHH410 pKa = 6.67KK411 pKa = 9.96NCEE414 pKa = 4.08QVSGYY419 pKa = 11.09AGTNTGIGTPGVSSSVINITGPGAWFHH446 pKa = 6.46TGLKK450 pKa = 10.08QGTPPQEE457 pKa = 4.55ANAKK461 pKa = 9.11MILIPIDD468 pKa = 4.79FDD470 pKa = 4.52TNWDD474 pKa = 3.44VTPPVV479 pKa = 3.38
Molecular weight: 51.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1505 |
479 |
1026 |
752.5 |
83.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.106 ± 0.203 | 0.93 ± 0.06 |
4.585 ± 0.227 | 5.98 ± 1.072 |
4.518 ± 0.04 | 6.777 ± 1.393 |
2.126 ± 0.243 | 4.983 ± 0.237 |
3.256 ± 0.378 | 9.568 ± 1.314 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.661 ± 0.116 | 3.455 ± 0.939 |
7.973 ± 1.244 | 3.721 ± 0.242 |
7.176 ± 1.041 | 8.372 ± 0.989 |
5.581 ± 1.252 | 6.512 ± 0.466 |
1.794 ± 0.288 | 2.924 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
