
Clostridium sp. chh4-2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; unclassified Clostridium
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
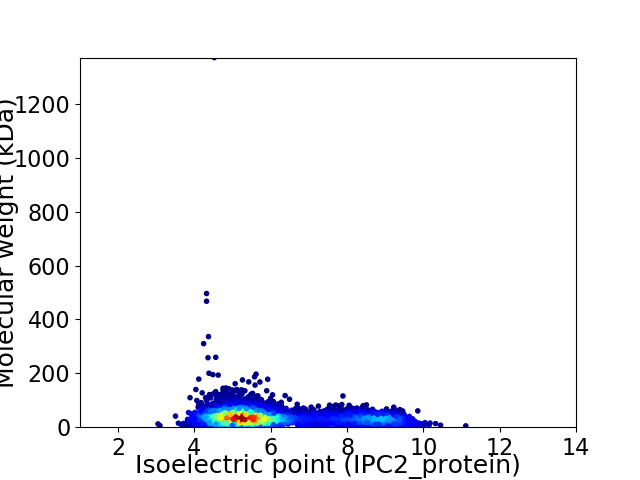
Virtual 2D-PAGE plot for 5111 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K2TUR2|A0A2K2TUR2_9CLOT Exopolyphosphatase OS=Clostridium sp. chh4-2 OX=2067550 GN=C0033_11435 PE=4 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84GMAAALAAMLTIGSALTGCGGGGTGGTTAASTAAPTEE41 pKa = 4.35TAASQAEE48 pKa = 4.34TSVGEE53 pKa = 4.23TNAPAKK59 pKa = 9.08EE60 pKa = 3.99LNYY63 pKa = 10.16ISTFDD68 pKa = 3.3VAGAGDD74 pKa = 3.59VALDD78 pKa = 3.92VMITSLGDD86 pKa = 3.52SDD88 pKa = 3.79GGPYY92 pKa = 10.3LRR94 pKa = 11.84GVMDD98 pKa = 6.0DD99 pKa = 3.81YY100 pKa = 11.32MAMYY104 pKa = 10.05PNVKK108 pKa = 9.85LNPVEE113 pKa = 5.23CSMNDD118 pKa = 3.25LYY120 pKa = 9.27TTLITQATAGTLPDD134 pKa = 3.67IFTMTEE140 pKa = 3.87AYY142 pKa = 9.76SANCLEE148 pKa = 4.71MGMSVDD154 pKa = 5.27NMAEE158 pKa = 4.04LLGDD162 pKa = 3.87EE163 pKa = 4.43YY164 pKa = 11.72LNGLMEE170 pKa = 4.51AAVDD174 pKa = 3.63NATVDD179 pKa = 3.43GTLVYY184 pKa = 9.94MPWQNNITAMVYY196 pKa = 10.28RR197 pKa = 11.84KK198 pKa = 10.27DD199 pKa = 3.38LFEE202 pKa = 4.49EE203 pKa = 4.28KK204 pKa = 10.44GIEE207 pKa = 4.49IPKK210 pKa = 8.75TWDD213 pKa = 3.13EE214 pKa = 4.1FLEE217 pKa = 4.31AAKK220 pKa = 9.63TLTEE224 pKa = 4.84DD225 pKa = 4.36LDD227 pKa = 5.34GDD229 pKa = 4.27GKK231 pKa = 8.89TDD233 pKa = 3.07RR234 pKa = 11.84YY235 pKa = 10.15GAAFAGTRR243 pKa = 11.84NDD245 pKa = 3.38SAEE248 pKa = 4.05SRR250 pKa = 11.84FQTFALTYY258 pKa = 9.76GCDD261 pKa = 3.37FVTDD265 pKa = 3.79NGDD268 pKa = 3.41GTFTSGFGTEE278 pKa = 4.04EE279 pKa = 4.2FKK281 pKa = 11.38NAMTSFVNMAANEE294 pKa = 4.32GVTPPGFIEE303 pKa = 4.06TGYY306 pKa = 10.39SEE308 pKa = 5.71AYY310 pKa = 9.41TMIAADD316 pKa = 3.8QACMFFSASNVLGGIYY332 pKa = 10.02NANPDD337 pKa = 3.74MKK339 pKa = 10.65GKK341 pKa = 9.22MGSFPMPTAEE351 pKa = 4.35GVDD354 pKa = 3.81PVTSFSSVGMTISNTCKK371 pKa = 10.24NPEE374 pKa = 3.79VAADD378 pKa = 3.56FLKK381 pKa = 11.29YY382 pKa = 7.52MTSVDD387 pKa = 5.12NSVTWNQATCRR398 pKa = 11.84LPVVKK403 pKa = 10.38DD404 pKa = 3.35ALTAICEE411 pKa = 3.99ADD413 pKa = 3.18EE414 pKa = 4.96AYY416 pKa = 10.79SGFADD421 pKa = 3.86ASDD424 pKa = 3.68SAVIYY429 pKa = 9.28PAFAGLAEE437 pKa = 4.46LRR439 pKa = 11.84DD440 pKa = 3.8ACGEE444 pKa = 3.99CWQTVVAEE452 pKa = 4.46GASIDD457 pKa = 4.03DD458 pKa = 3.93AVAAAAAKK466 pKa = 10.64AEE468 pKa = 4.38DD469 pKa = 3.75VAKK472 pKa = 10.43SYY474 pKa = 11.59SKK476 pKa = 11.32
MM1 pKa = 7.78RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84GMAAALAAMLTIGSALTGCGGGGTGGTTAASTAAPTEE41 pKa = 4.35TAASQAEE48 pKa = 4.34TSVGEE53 pKa = 4.23TNAPAKK59 pKa = 9.08EE60 pKa = 3.99LNYY63 pKa = 10.16ISTFDD68 pKa = 3.3VAGAGDD74 pKa = 3.59VALDD78 pKa = 3.92VMITSLGDD86 pKa = 3.52SDD88 pKa = 3.79GGPYY92 pKa = 10.3LRR94 pKa = 11.84GVMDD98 pKa = 6.0DD99 pKa = 3.81YY100 pKa = 11.32MAMYY104 pKa = 10.05PNVKK108 pKa = 9.85LNPVEE113 pKa = 5.23CSMNDD118 pKa = 3.25LYY120 pKa = 9.27TTLITQATAGTLPDD134 pKa = 3.67IFTMTEE140 pKa = 3.87AYY142 pKa = 9.76SANCLEE148 pKa = 4.71MGMSVDD154 pKa = 5.27NMAEE158 pKa = 4.04LLGDD162 pKa = 3.87EE163 pKa = 4.43YY164 pKa = 11.72LNGLMEE170 pKa = 4.51AAVDD174 pKa = 3.63NATVDD179 pKa = 3.43GTLVYY184 pKa = 9.94MPWQNNITAMVYY196 pKa = 10.28RR197 pKa = 11.84KK198 pKa = 10.27DD199 pKa = 3.38LFEE202 pKa = 4.49EE203 pKa = 4.28KK204 pKa = 10.44GIEE207 pKa = 4.49IPKK210 pKa = 8.75TWDD213 pKa = 3.13EE214 pKa = 4.1FLEE217 pKa = 4.31AAKK220 pKa = 9.63TLTEE224 pKa = 4.84DD225 pKa = 4.36LDD227 pKa = 5.34GDD229 pKa = 4.27GKK231 pKa = 8.89TDD233 pKa = 3.07RR234 pKa = 11.84YY235 pKa = 10.15GAAFAGTRR243 pKa = 11.84NDD245 pKa = 3.38SAEE248 pKa = 4.05SRR250 pKa = 11.84FQTFALTYY258 pKa = 9.76GCDD261 pKa = 3.37FVTDD265 pKa = 3.79NGDD268 pKa = 3.41GTFTSGFGTEE278 pKa = 4.04EE279 pKa = 4.2FKK281 pKa = 11.38NAMTSFVNMAANEE294 pKa = 4.32GVTPPGFIEE303 pKa = 4.06TGYY306 pKa = 10.39SEE308 pKa = 5.71AYY310 pKa = 9.41TMIAADD316 pKa = 3.8QACMFFSASNVLGGIYY332 pKa = 10.02NANPDD337 pKa = 3.74MKK339 pKa = 10.65GKK341 pKa = 9.22MGSFPMPTAEE351 pKa = 4.35GVDD354 pKa = 3.81PVTSFSSVGMTISNTCKK371 pKa = 10.24NPEE374 pKa = 3.79VAADD378 pKa = 3.56FLKK381 pKa = 11.29YY382 pKa = 7.52MTSVDD387 pKa = 5.12NSVTWNQATCRR398 pKa = 11.84LPVVKK403 pKa = 10.38DD404 pKa = 3.35ALTAICEE411 pKa = 3.99ADD413 pKa = 3.18EE414 pKa = 4.96AYY416 pKa = 10.79SGFADD421 pKa = 3.86ASDD424 pKa = 3.68SAVIYY429 pKa = 9.28PAFAGLAEE437 pKa = 4.46LRR439 pKa = 11.84DD440 pKa = 3.8ACGEE444 pKa = 3.99CWQTVVAEE452 pKa = 4.46GASIDD457 pKa = 4.03DD458 pKa = 3.93AVAAAAAKK466 pKa = 10.64AEE468 pKa = 4.38DD469 pKa = 3.75VAKK472 pKa = 10.43SYY474 pKa = 11.59SKK476 pKa = 11.32
Molecular weight: 50.04 kDa
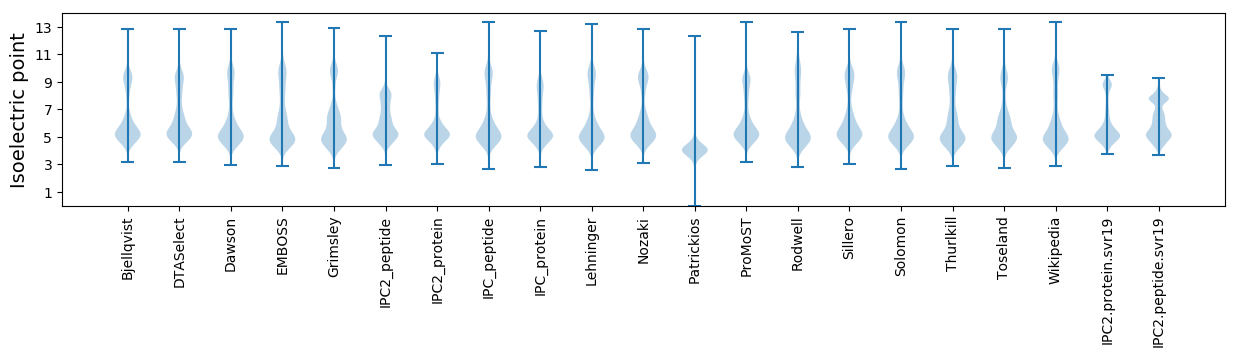
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K2TV41|A0A2K2TV41_9CLOT 1-acyl-sn-glycerol-3-phosphate acyltransferase OS=Clostridium sp. chh4-2 OX=2067550 GN=C0033_11420 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.63NRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.89VHH16 pKa = 5.88GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 9.06VIASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.4GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.67KK2 pKa = 8.73MTFQPKK8 pKa = 8.63NRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.89VHH16 pKa = 5.88GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 9.06VIASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.4GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 4.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1759323 |
27 |
12064 |
344.2 |
38.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.475 ± 0.04 | 1.545 ± 0.017 |
5.551 ± 0.039 | 7.359 ± 0.04 |
4.216 ± 0.029 | 7.36 ± 0.032 |
1.734 ± 0.017 | 7.312 ± 0.036 |
6.383 ± 0.025 | 9.115 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.284 ± 0.022 | 4.251 ± 0.026 |
3.61 ± 0.021 | 3.212 ± 0.021 |
4.291 ± 0.026 | 5.987 ± 0.026 |
5.257 ± 0.036 | 6.8 ± 0.03 |
1.025 ± 0.013 | 4.233 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
