
Salana multivorans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Beutenbergiaceae; Salana
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
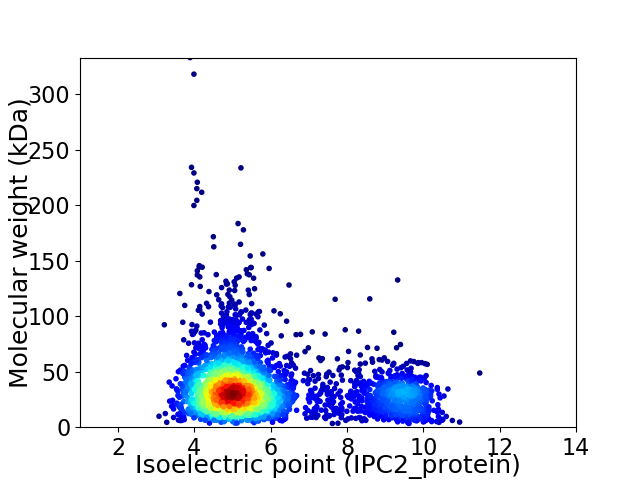
Virtual 2D-PAGE plot for 3332 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N2DBE7|A0A3N2DBE7_9MICO Exoribonuclease R OS=Salana multivorans OX=120377 GN=EDD28_1712 PE=4 SV=1
MM1 pKa = 7.23RR2 pKa = 11.84TSSSRR7 pKa = 11.84VLAAIVALGVCLGLTACVPGLIDD30 pKa = 4.5PFAGGNGSSTDD41 pKa = 3.66PSDD44 pKa = 5.55ADD46 pKa = 3.95DD47 pKa = 4.11TTDD50 pKa = 4.56PDD52 pKa = 4.39DD53 pKa = 4.87PSDD56 pKa = 3.53AAEE59 pKa = 4.46APVVLDD65 pKa = 4.65PGPSSTAIDD74 pKa = 3.72WRR76 pKa = 11.84PCDD79 pKa = 3.11HH80 pKa = 7.22HH81 pKa = 8.52ARR83 pKa = 11.84FDD85 pKa = 3.85CAEE88 pKa = 3.86VLLPLDD94 pKa = 4.13HH95 pKa = 7.51ADD97 pKa = 3.59PGGDD101 pKa = 3.13QVRR104 pKa = 11.84IALRR108 pKa = 11.84RR109 pKa = 11.84LPAGGPEE116 pKa = 3.96EE117 pKa = 4.44EE118 pKa = 4.49RR119 pKa = 11.84LGTLWINPGGPGEE132 pKa = 4.19SGIDD136 pKa = 3.62LVTMVGSFFGDD147 pKa = 3.53DD148 pKa = 3.14LLEE151 pKa = 5.2RR152 pKa = 11.84YY153 pKa = 9.68DD154 pKa = 3.93VVGFDD159 pKa = 3.76PRR161 pKa = 11.84GVGASTHH168 pKa = 5.09VVCLLDD174 pKa = 5.63GEE176 pKa = 5.57DD177 pKa = 3.78DD178 pKa = 4.39PSWVDD183 pKa = 4.98LDD185 pKa = 4.61LDD187 pKa = 4.14TPKK190 pKa = 11.09GLARR194 pKa = 11.84VEE196 pKa = 4.48SGFTALGEE204 pKa = 4.21LCRR207 pKa = 11.84EE208 pKa = 3.87RR209 pKa = 11.84SGALLDD215 pKa = 4.08HH216 pKa = 6.92VSTADD221 pKa = 3.4TVADD225 pKa = 4.46LEE227 pKa = 4.31ALRR230 pKa = 11.84ALTGEE235 pKa = 3.88QLNYY239 pKa = 10.54LGYY242 pKa = 10.64SYY244 pKa = 10.14GTLIGAVYY252 pKa = 10.84ADD254 pKa = 4.56TYY256 pKa = 9.17PEE258 pKa = 3.84RR259 pKa = 11.84VGRR262 pKa = 11.84MVLDD266 pKa = 4.0GAIDD270 pKa = 3.85PSVGYY275 pKa = 9.06TDD277 pKa = 5.48LQAQQTWTFQRR288 pKa = 11.84AFARR292 pKa = 11.84FVTTCRR298 pKa = 11.84EE299 pKa = 4.07DD300 pKa = 3.56ADD302 pKa = 4.62CPLDD306 pKa = 4.12ADD308 pKa = 3.8PRR310 pKa = 11.84AAEE313 pKa = 4.18TQVLDD318 pKa = 4.47WIDD321 pKa = 3.92EE322 pKa = 4.38LDD324 pKa = 3.85VSPVPSDD331 pKa = 4.02AEE333 pKa = 4.4DD334 pKa = 4.46GEE336 pKa = 4.58LTGQEE341 pKa = 4.14LVAALASALYY351 pKa = 10.34APWDD355 pKa = 3.4WDD357 pKa = 3.88VVVEE361 pKa = 4.39DD362 pKa = 4.83LADD365 pKa = 3.61AVADD369 pKa = 3.89SDD371 pKa = 4.44PDD373 pKa = 3.89GSALDD378 pKa = 3.82EE379 pKa = 4.98LQGEE383 pKa = 4.84WYY385 pKa = 10.34GGALNYY391 pKa = 9.11PFWFSAIDD399 pKa = 3.75CVDD402 pKa = 3.73YY403 pKa = 10.76PVGTFDD409 pKa = 4.67EE410 pKa = 5.0AVTHH414 pKa = 5.78ARR416 pKa = 11.84EE417 pKa = 3.95LAAGSPVFGAGSIAEE432 pKa = 4.36IQCATWPAKK441 pKa = 10.62ARR443 pKa = 11.84DD444 pKa = 3.55EE445 pKa = 4.12RR446 pKa = 11.84VPVSGTGAAPILVVGTLFDD465 pKa = 4.24PATPYY470 pKa = 10.95AWAVSLSRR478 pKa = 11.84QLEE481 pKa = 4.37SATLLTYY488 pKa = 10.61EE489 pKa = 4.56GDD491 pKa = 3.17GHH493 pKa = 6.92AVYY496 pKa = 10.36GGVSWCVDD504 pKa = 3.32GFVEE508 pKa = 6.25DD509 pKa = 4.1YY510 pKa = 10.88LIDD513 pKa = 4.13GAVPPPGMSCPSAFF527 pKa = 3.76
MM1 pKa = 7.23RR2 pKa = 11.84TSSSRR7 pKa = 11.84VLAAIVALGVCLGLTACVPGLIDD30 pKa = 4.5PFAGGNGSSTDD41 pKa = 3.66PSDD44 pKa = 5.55ADD46 pKa = 3.95DD47 pKa = 4.11TTDD50 pKa = 4.56PDD52 pKa = 4.39DD53 pKa = 4.87PSDD56 pKa = 3.53AAEE59 pKa = 4.46APVVLDD65 pKa = 4.65PGPSSTAIDD74 pKa = 3.72WRR76 pKa = 11.84PCDD79 pKa = 3.11HH80 pKa = 7.22HH81 pKa = 8.52ARR83 pKa = 11.84FDD85 pKa = 3.85CAEE88 pKa = 3.86VLLPLDD94 pKa = 4.13HH95 pKa = 7.51ADD97 pKa = 3.59PGGDD101 pKa = 3.13QVRR104 pKa = 11.84IALRR108 pKa = 11.84RR109 pKa = 11.84LPAGGPEE116 pKa = 3.96EE117 pKa = 4.44EE118 pKa = 4.49RR119 pKa = 11.84LGTLWINPGGPGEE132 pKa = 4.19SGIDD136 pKa = 3.62LVTMVGSFFGDD147 pKa = 3.53DD148 pKa = 3.14LLEE151 pKa = 5.2RR152 pKa = 11.84YY153 pKa = 9.68DD154 pKa = 3.93VVGFDD159 pKa = 3.76PRR161 pKa = 11.84GVGASTHH168 pKa = 5.09VVCLLDD174 pKa = 5.63GEE176 pKa = 5.57DD177 pKa = 3.78DD178 pKa = 4.39PSWVDD183 pKa = 4.98LDD185 pKa = 4.61LDD187 pKa = 4.14TPKK190 pKa = 11.09GLARR194 pKa = 11.84VEE196 pKa = 4.48SGFTALGEE204 pKa = 4.21LCRR207 pKa = 11.84EE208 pKa = 3.87RR209 pKa = 11.84SGALLDD215 pKa = 4.08HH216 pKa = 6.92VSTADD221 pKa = 3.4TVADD225 pKa = 4.46LEE227 pKa = 4.31ALRR230 pKa = 11.84ALTGEE235 pKa = 3.88QLNYY239 pKa = 10.54LGYY242 pKa = 10.64SYY244 pKa = 10.14GTLIGAVYY252 pKa = 10.84ADD254 pKa = 4.56TYY256 pKa = 9.17PEE258 pKa = 3.84RR259 pKa = 11.84VGRR262 pKa = 11.84MVLDD266 pKa = 4.0GAIDD270 pKa = 3.85PSVGYY275 pKa = 9.06TDD277 pKa = 5.48LQAQQTWTFQRR288 pKa = 11.84AFARR292 pKa = 11.84FVTTCRR298 pKa = 11.84EE299 pKa = 4.07DD300 pKa = 3.56ADD302 pKa = 4.62CPLDD306 pKa = 4.12ADD308 pKa = 3.8PRR310 pKa = 11.84AAEE313 pKa = 4.18TQVLDD318 pKa = 4.47WIDD321 pKa = 3.92EE322 pKa = 4.38LDD324 pKa = 3.85VSPVPSDD331 pKa = 4.02AEE333 pKa = 4.4DD334 pKa = 4.46GEE336 pKa = 4.58LTGQEE341 pKa = 4.14LVAALASALYY351 pKa = 10.34APWDD355 pKa = 3.4WDD357 pKa = 3.88VVVEE361 pKa = 4.39DD362 pKa = 4.83LADD365 pKa = 3.61AVADD369 pKa = 3.89SDD371 pKa = 4.44PDD373 pKa = 3.89GSALDD378 pKa = 3.82EE379 pKa = 4.98LQGEE383 pKa = 4.84WYY385 pKa = 10.34GGALNYY391 pKa = 9.11PFWFSAIDD399 pKa = 3.75CVDD402 pKa = 3.73YY403 pKa = 10.76PVGTFDD409 pKa = 4.67EE410 pKa = 5.0AVTHH414 pKa = 5.78ARR416 pKa = 11.84EE417 pKa = 3.95LAAGSPVFGAGSIAEE432 pKa = 4.36IQCATWPAKK441 pKa = 10.62ARR443 pKa = 11.84DD444 pKa = 3.55EE445 pKa = 4.12RR446 pKa = 11.84VPVSGTGAAPILVVGTLFDD465 pKa = 4.24PATPYY470 pKa = 10.95AWAVSLSRR478 pKa = 11.84QLEE481 pKa = 4.37SATLLTYY488 pKa = 10.61EE489 pKa = 4.56GDD491 pKa = 3.17GHH493 pKa = 6.92AVYY496 pKa = 10.36GGVSWCVDD504 pKa = 3.32GFVEE508 pKa = 6.25DD509 pKa = 4.1YY510 pKa = 10.88LIDD513 pKa = 4.13GAVPPPGMSCPSAFF527 pKa = 3.76
Molecular weight: 55.66 kDa
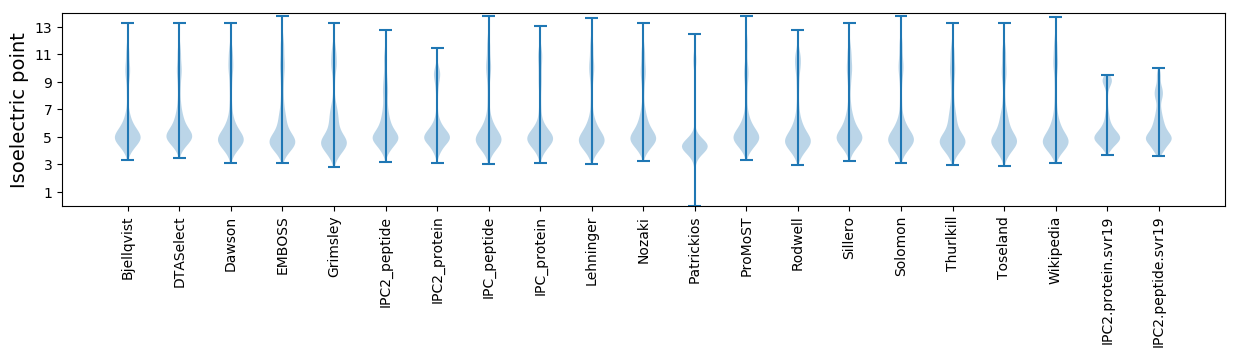
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N2D223|A0A3N2D223_9MICO 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase OS=Salana multivorans OX=120377 GN=hisA PE=3 SV=1
MM1 pKa = 6.12VTSVLVVPTTTGRR14 pKa = 11.84RR15 pKa = 11.84VARR18 pKa = 11.84VTVPRR23 pKa = 11.84VPTTTVRR30 pKa = 11.84SVRR33 pKa = 11.84TRR35 pKa = 11.84SAVRR39 pKa = 11.84VVTVLTVRR47 pKa = 11.84SVPTRR52 pKa = 11.84TVVLAARR59 pKa = 11.84VTVPRR64 pKa = 11.84VPTTTVRR71 pKa = 11.84SVRR74 pKa = 11.84TRR76 pKa = 11.84TVVRR80 pKa = 11.84VVTVLTVRR88 pKa = 11.84SVPTRR93 pKa = 11.84TVVLAARR100 pKa = 11.84VTGPRR105 pKa = 11.84VPTTTVRR112 pKa = 11.84SVRR115 pKa = 11.84TRR117 pKa = 11.84TVVRR121 pKa = 11.84VVTVLTVRR129 pKa = 11.84SVPTTTAVPVARR141 pKa = 11.84VTGPRR146 pKa = 11.84VPTTTVRR153 pKa = 11.84SVRR156 pKa = 11.84TRR158 pKa = 11.84TVVRR162 pKa = 11.84AARR165 pKa = 11.84VTGPRR170 pKa = 11.84VPTTTGPSVRR180 pKa = 11.84TRR182 pKa = 11.84TAVRR186 pKa = 11.84VATVLTGPRR195 pKa = 11.84VPTTTAVRR203 pKa = 11.84AVRR206 pKa = 11.84AKK208 pKa = 9.27TGRR211 pKa = 11.84AARR214 pKa = 11.84TVMTAPSVPMRR225 pKa = 11.84TVARR229 pKa = 11.84VATVTSVPVVLTTTGPSVPTRR250 pKa = 11.84TVVRR254 pKa = 11.84VVRR257 pKa = 11.84ASVPRR262 pKa = 11.84VPMVMTVRR270 pKa = 11.84VARR273 pKa = 11.84TATTVVRR280 pKa = 11.84VVTVLTDD287 pKa = 3.56PRR289 pKa = 11.84APTVMTVRR297 pKa = 11.84SVPTTTAALVVTVTSVPVVPTVMTVRR323 pKa = 11.84SVRR326 pKa = 11.84TRR328 pKa = 11.84SAALVVRR335 pKa = 11.84VSGPHH340 pKa = 5.77VPTAMIVRR348 pKa = 11.84SVLTTTAARR357 pKa = 11.84VAAVTSAPVVRR368 pKa = 11.84TATTVRR374 pKa = 11.84SVPTTTAAHH383 pKa = 5.98VVTAMIVRR391 pKa = 11.84VAPTATTGGLAVTATTAAGARR412 pKa = 11.84PTRR415 pKa = 11.84ARR417 pKa = 11.84LVRR420 pKa = 11.84STSGVARR427 pKa = 11.84TATSARR433 pKa = 11.84ARR435 pKa = 11.84TVAPSASTTRR445 pKa = 11.84SSPRR449 pKa = 11.84RR450 pKa = 11.84SSSRR454 pKa = 11.84CSRR457 pKa = 11.84ARR459 pKa = 11.84LARR462 pKa = 11.84RR463 pKa = 11.84CAAA466 pKa = 4.02
MM1 pKa = 6.12VTSVLVVPTTTGRR14 pKa = 11.84RR15 pKa = 11.84VARR18 pKa = 11.84VTVPRR23 pKa = 11.84VPTTTVRR30 pKa = 11.84SVRR33 pKa = 11.84TRR35 pKa = 11.84SAVRR39 pKa = 11.84VVTVLTVRR47 pKa = 11.84SVPTRR52 pKa = 11.84TVVLAARR59 pKa = 11.84VTVPRR64 pKa = 11.84VPTTTVRR71 pKa = 11.84SVRR74 pKa = 11.84TRR76 pKa = 11.84TVVRR80 pKa = 11.84VVTVLTVRR88 pKa = 11.84SVPTRR93 pKa = 11.84TVVLAARR100 pKa = 11.84VTGPRR105 pKa = 11.84VPTTTVRR112 pKa = 11.84SVRR115 pKa = 11.84TRR117 pKa = 11.84TVVRR121 pKa = 11.84VVTVLTVRR129 pKa = 11.84SVPTTTAVPVARR141 pKa = 11.84VTGPRR146 pKa = 11.84VPTTTVRR153 pKa = 11.84SVRR156 pKa = 11.84TRR158 pKa = 11.84TVVRR162 pKa = 11.84AARR165 pKa = 11.84VTGPRR170 pKa = 11.84VPTTTGPSVRR180 pKa = 11.84TRR182 pKa = 11.84TAVRR186 pKa = 11.84VATVLTGPRR195 pKa = 11.84VPTTTAVRR203 pKa = 11.84AVRR206 pKa = 11.84AKK208 pKa = 9.27TGRR211 pKa = 11.84AARR214 pKa = 11.84TVMTAPSVPMRR225 pKa = 11.84TVARR229 pKa = 11.84VATVTSVPVVLTTTGPSVPTRR250 pKa = 11.84TVVRR254 pKa = 11.84VVRR257 pKa = 11.84ASVPRR262 pKa = 11.84VPMVMTVRR270 pKa = 11.84VARR273 pKa = 11.84TATTVVRR280 pKa = 11.84VVTVLTDD287 pKa = 3.56PRR289 pKa = 11.84APTVMTVRR297 pKa = 11.84SVPTTTAALVVTVTSVPVVPTVMTVRR323 pKa = 11.84SVRR326 pKa = 11.84TRR328 pKa = 11.84SAALVVRR335 pKa = 11.84VSGPHH340 pKa = 5.77VPTAMIVRR348 pKa = 11.84SVLTTTAARR357 pKa = 11.84VAAVTSAPVVRR368 pKa = 11.84TATTVRR374 pKa = 11.84SVPTTTAAHH383 pKa = 5.98VVTAMIVRR391 pKa = 11.84VAPTATTGGLAVTATTAAGARR412 pKa = 11.84PTRR415 pKa = 11.84ARR417 pKa = 11.84LVRR420 pKa = 11.84STSGVARR427 pKa = 11.84TATSARR433 pKa = 11.84ARR435 pKa = 11.84TVAPSASTTRR445 pKa = 11.84SSPRR449 pKa = 11.84RR450 pKa = 11.84SSSRR454 pKa = 11.84CSRR457 pKa = 11.84ARR459 pKa = 11.84LARR462 pKa = 11.84RR463 pKa = 11.84CAAA466 pKa = 4.02
Molecular weight: 48.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1173099 |
30 |
3301 |
352.1 |
37.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.021 ± 0.059 | 0.55 ± 0.011 |
6.482 ± 0.036 | 5.74 ± 0.038 |
2.533 ± 0.023 | 9.605 ± 0.044 |
1.896 ± 0.02 | 3.362 ± 0.027 |
1.299 ± 0.024 | 10.239 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.584 ± 0.015 | 1.603 ± 0.022 |
6.039 ± 0.035 | 2.424 ± 0.022 |
7.722 ± 0.058 | 5.33 ± 0.031 |
6.385 ± 0.059 | 9.633 ± 0.041 |
1.644 ± 0.021 | 1.91 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
