
Monosiga brevicollis (Choanoflagellate)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Choanoflagellata; Craspedida; Salpingoecidae; Monosiga
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
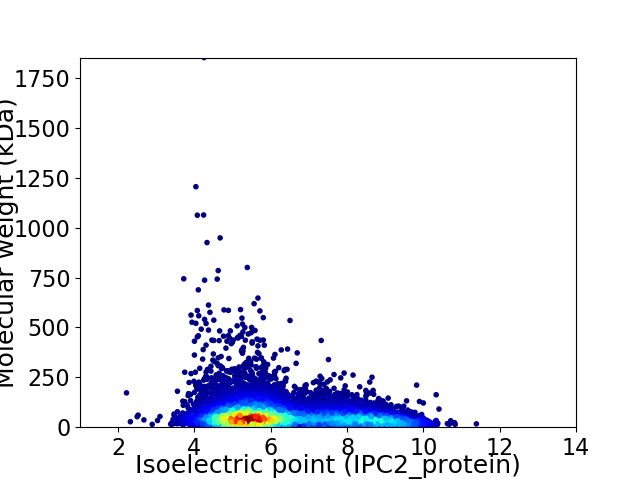
Virtual 2D-PAGE plot for 9188 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A9UQI2|A9UQI2_MONBE Uncharacterized protein OS=Monosiga brevicollis OX=81824 GN=21077 PE=3 SV=1
MM1 pKa = 7.6GLRR4 pKa = 11.84RR5 pKa = 11.84SWAGFCLGVGLVLVLVACDD24 pKa = 3.15VGAGVEE30 pKa = 4.3EE31 pKa = 4.54PSLAFDD37 pKa = 3.75EE38 pKa = 4.98NGTLFINATNRR49 pKa = 11.84VVVNGVDD56 pKa = 3.76VQALAMAFSAASSSLDD72 pKa = 3.59ALIEE76 pKa = 4.02LQRR79 pKa = 11.84EE80 pKa = 4.15LNPPQFEE87 pKa = 3.94QHH89 pKa = 6.55AISATMDD96 pKa = 3.67GSRR99 pKa = 11.84SACAADD105 pKa = 3.89LDD107 pKa = 4.46GDD109 pKa = 4.05GFLDD113 pKa = 3.85VLSTNYY119 pKa = 9.79LQHH122 pKa = 6.18QISWYY127 pKa = 9.77RR128 pKa = 11.84NNGDD132 pKa = 3.95GSFSDD137 pKa = 4.22QIVITNTSQSVWYY150 pKa = 7.26ATPADD155 pKa = 4.02LSGDD159 pKa = 3.73GQLDD163 pKa = 3.63IVSVWHH169 pKa = 6.74NDD171 pKa = 3.05MIAWYY176 pKa = 10.15LNTGNGAFSPEE187 pKa = 3.87PMIITTDD194 pKa = 3.3VKK196 pKa = 10.7AGRR199 pKa = 11.84SVFATDD205 pKa = 4.33FNNDD209 pKa = 2.64GYY211 pKa = 9.46TDD213 pKa = 3.8VVSASMDD220 pKa = 3.63DD221 pKa = 6.07DD222 pKa = 4.65KK223 pKa = 11.49IAWYY227 pKa = 10.12RR228 pKa = 11.84NNGNSTFSAQIIISTDD244 pKa = 2.97AVEE247 pKa = 4.35ALSVYY252 pKa = 10.64AADD255 pKa = 4.52LNNDD259 pKa = 3.11GFADD263 pKa = 3.85VLSASPGDD271 pKa = 5.26DD272 pKa = 2.67KK273 pKa = 11.72VAWYY277 pKa = 10.19RR278 pKa = 11.84NNGNGTFTNQIVITTSADD296 pKa = 3.5GPVSVYY302 pKa = 10.82AADD305 pKa = 4.21LDD307 pKa = 4.52GDD309 pKa = 3.83GHH311 pKa = 8.82LDD313 pKa = 3.6VLSASYY319 pKa = 11.01LDD321 pKa = 5.87NKK323 pKa = 10.04IAWCRR328 pKa = 11.84NNGNGTFSEE337 pKa = 4.17QLVISDD343 pKa = 4.55AADD346 pKa = 3.25MAYY349 pKa = 10.22SVYY352 pKa = 10.83AADD355 pKa = 5.19LDD357 pKa = 4.29NDD359 pKa = 3.54GHH361 pKa = 8.65LDD363 pKa = 3.66VLSASYY369 pKa = 11.27YY370 pKa = 9.65DD371 pKa = 6.19DD372 pKa = 5.16KK373 pKa = 11.27IAWYY377 pKa = 9.6RR378 pKa = 11.84NKK380 pKa = 10.92GNGTFSDD387 pKa = 3.49QLIITEE393 pKa = 4.42AANGAYY399 pKa = 9.98SVFAADD405 pKa = 4.84LDD407 pKa = 4.32NNGDD411 pKa = 3.49IDD413 pKa = 4.09ILSASYY419 pKa = 10.69LGDD422 pKa = 4.37GIAWYY427 pKa = 10.71RR428 pKa = 11.84NDD430 pKa = 4.54DD431 pKa = 3.74PVAHH435 pKa = 6.97FFRR438 pKa = 11.84STT440 pKa = 2.86
MM1 pKa = 7.6GLRR4 pKa = 11.84RR5 pKa = 11.84SWAGFCLGVGLVLVLVACDD24 pKa = 3.15VGAGVEE30 pKa = 4.3EE31 pKa = 4.54PSLAFDD37 pKa = 3.75EE38 pKa = 4.98NGTLFINATNRR49 pKa = 11.84VVVNGVDD56 pKa = 3.76VQALAMAFSAASSSLDD72 pKa = 3.59ALIEE76 pKa = 4.02LQRR79 pKa = 11.84EE80 pKa = 4.15LNPPQFEE87 pKa = 3.94QHH89 pKa = 6.55AISATMDD96 pKa = 3.67GSRR99 pKa = 11.84SACAADD105 pKa = 3.89LDD107 pKa = 4.46GDD109 pKa = 4.05GFLDD113 pKa = 3.85VLSTNYY119 pKa = 9.79LQHH122 pKa = 6.18QISWYY127 pKa = 9.77RR128 pKa = 11.84NNGDD132 pKa = 3.95GSFSDD137 pKa = 4.22QIVITNTSQSVWYY150 pKa = 7.26ATPADD155 pKa = 4.02LSGDD159 pKa = 3.73GQLDD163 pKa = 3.63IVSVWHH169 pKa = 6.74NDD171 pKa = 3.05MIAWYY176 pKa = 10.15LNTGNGAFSPEE187 pKa = 3.87PMIITTDD194 pKa = 3.3VKK196 pKa = 10.7AGRR199 pKa = 11.84SVFATDD205 pKa = 4.33FNNDD209 pKa = 2.64GYY211 pKa = 9.46TDD213 pKa = 3.8VVSASMDD220 pKa = 3.63DD221 pKa = 6.07DD222 pKa = 4.65KK223 pKa = 11.49IAWYY227 pKa = 10.12RR228 pKa = 11.84NNGNSTFSAQIIISTDD244 pKa = 2.97AVEE247 pKa = 4.35ALSVYY252 pKa = 10.64AADD255 pKa = 4.52LNNDD259 pKa = 3.11GFADD263 pKa = 3.85VLSASPGDD271 pKa = 5.26DD272 pKa = 2.67KK273 pKa = 11.72VAWYY277 pKa = 10.19RR278 pKa = 11.84NNGNGTFTNQIVITTSADD296 pKa = 3.5GPVSVYY302 pKa = 10.82AADD305 pKa = 4.21LDD307 pKa = 4.52GDD309 pKa = 3.83GHH311 pKa = 8.82LDD313 pKa = 3.6VLSASYY319 pKa = 11.01LDD321 pKa = 5.87NKK323 pKa = 10.04IAWCRR328 pKa = 11.84NNGNGTFSEE337 pKa = 4.17QLVISDD343 pKa = 4.55AADD346 pKa = 3.25MAYY349 pKa = 10.22SVYY352 pKa = 10.83AADD355 pKa = 5.19LDD357 pKa = 4.29NDD359 pKa = 3.54GHH361 pKa = 8.65LDD363 pKa = 3.66VLSASYY369 pKa = 11.27YY370 pKa = 9.65DD371 pKa = 6.19DD372 pKa = 5.16KK373 pKa = 11.27IAWYY377 pKa = 9.6RR378 pKa = 11.84NKK380 pKa = 10.92GNGTFSDD387 pKa = 3.49QLIITEE393 pKa = 4.42AANGAYY399 pKa = 9.98SVFAADD405 pKa = 4.84LDD407 pKa = 4.32NNGDD411 pKa = 3.49IDD413 pKa = 4.09ILSASYY419 pKa = 10.69LGDD422 pKa = 4.37GIAWYY427 pKa = 10.71RR428 pKa = 11.84NDD430 pKa = 4.54DD431 pKa = 3.74PVAHH435 pKa = 6.97FFRR438 pKa = 11.84STT440 pKa = 2.86
Molecular weight: 47.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9V395|A9V395_MONBE Predicted protein OS=Monosiga brevicollis OX=81824 GN=9474 PE=4 SV=1
MM1 pKa = 7.54ASKK4 pKa = 10.31AANAPAARR12 pKa = 11.84VPRR15 pKa = 11.84GAIVPRR21 pKa = 11.84LASAPTARR29 pKa = 11.84AAKK32 pKa = 9.67RR33 pKa = 11.84VTASAVATVLPLAARR48 pKa = 11.84ATARR52 pKa = 11.84TASAARR58 pKa = 11.84RR59 pKa = 11.84AATAPAAVTAPAATSATAPTASAARR84 pKa = 11.84RR85 pKa = 11.84ALRR88 pKa = 11.84PLPLAVALAANVATPVLAATSASAAKK114 pKa = 10.01IIVGTKK120 pKa = 7.58TLSVLSVRR128 pKa = 11.84LLWFTPSGTKK138 pKa = 7.39THH140 pKa = 6.95HH141 pKa = 6.35ARR143 pKa = 11.84TLASSSLFRR152 pKa = 11.84KK153 pKa = 7.88QTLFGIIQFWFGPP166 pKa = 3.74
MM1 pKa = 7.54ASKK4 pKa = 10.31AANAPAARR12 pKa = 11.84VPRR15 pKa = 11.84GAIVPRR21 pKa = 11.84LASAPTARR29 pKa = 11.84AAKK32 pKa = 9.67RR33 pKa = 11.84VTASAVATVLPLAARR48 pKa = 11.84ATARR52 pKa = 11.84TASAARR58 pKa = 11.84RR59 pKa = 11.84AATAPAAVTAPAATSATAPTASAARR84 pKa = 11.84RR85 pKa = 11.84ALRR88 pKa = 11.84PLPLAVALAANVATPVLAATSASAAKK114 pKa = 10.01IIVGTKK120 pKa = 7.58TLSVLSVRR128 pKa = 11.84LLWFTPSGTKK138 pKa = 7.39THH140 pKa = 6.95HH141 pKa = 6.35ARR143 pKa = 11.84TLASSSLFRR152 pKa = 11.84KK153 pKa = 7.88QTLFGIIQFWFGPP166 pKa = 3.74
Molecular weight: 16.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5511963 |
49 |
17829 |
599.9 |
65.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.643 ± 0.033 | 1.819 ± 0.019 |
5.836 ± 0.021 | 5.765 ± 0.029 |
3.335 ± 0.014 | 6.068 ± 0.03 |
2.801 ± 0.016 | 3.686 ± 0.016 |
3.485 ± 0.03 | 10.149 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.178 ± 0.013 | 3.401 ± 0.018 |
5.537 ± 0.026 | 4.658 ± 0.022 |
6.321 ± 0.028 | 7.752 ± 0.031 |
6.259 ± 0.036 | 6.787 ± 0.035 |
1.164 ± 0.009 | 2.355 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
