
Halobaculum gomorrense
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halobaculum
Average proteome isoelectric point is 4.91
Get precalculated fractions of proteins
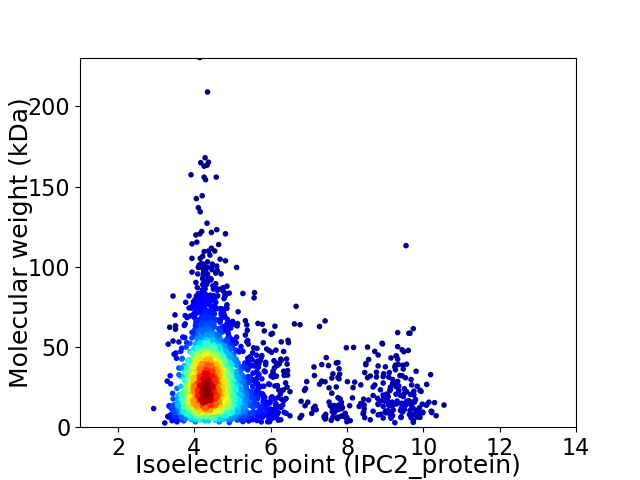
Virtual 2D-PAGE plot for 3149 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
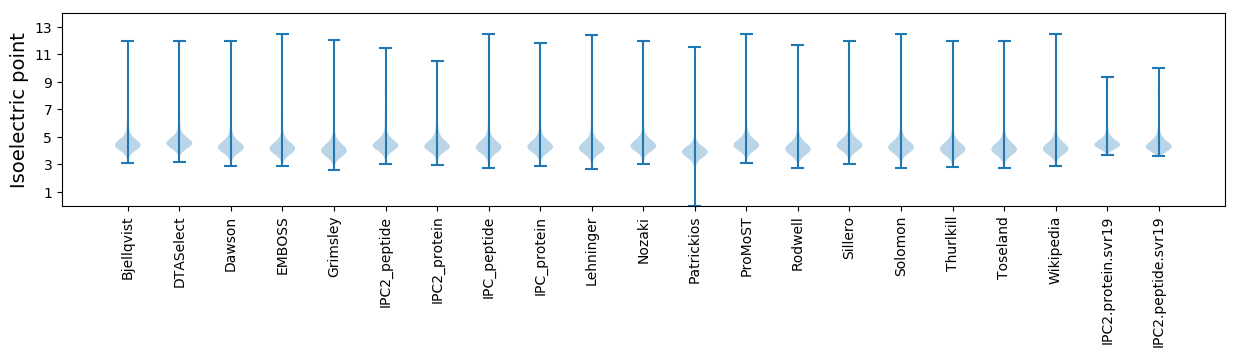
* You can choose from 21 different methods for calculating isoelectric point
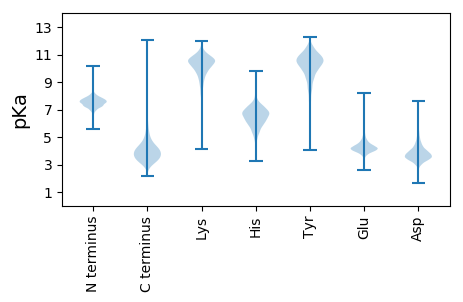
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5QKD5|A0A1M5QKD5_9EURY 3-oxoacyl-[acyl-carrier protein] reductase OS=Halobaculum gomorrense OX=43928 GN=SAMN05443636_1933 PE=4 SV=1
MM1 pKa = 7.59GIDD4 pKa = 4.45PNFDD8 pKa = 3.82DD9 pKa = 4.06NRR11 pKa = 11.84EE12 pKa = 4.18HH13 pKa = 8.17VGEE16 pKa = 4.12EE17 pKa = 3.65NGLDD21 pKa = 3.13VWGPVDD27 pKa = 3.89PPEE30 pKa = 4.39KK31 pKa = 10.5LGIHH35 pKa = 6.29GAHH38 pKa = 6.12VAVDD42 pKa = 3.77YY43 pKa = 10.01DD44 pKa = 3.37ICIADD49 pKa = 4.36GACLEE54 pKa = 4.2NCPVDD59 pKa = 3.64VFDD62 pKa = 5.23WVDD65 pKa = 3.63TPDD68 pKa = 3.53HH69 pKa = 6.97PEE71 pKa = 3.86SEE73 pKa = 4.48KK74 pKa = 10.81KK75 pKa = 10.45VEE77 pKa = 4.04PTRR80 pKa = 11.84EE81 pKa = 4.05DD82 pKa = 3.22QCIDD86 pKa = 3.44CMLCVDD92 pKa = 4.88ICPVDD97 pKa = 5.13AIDD100 pKa = 3.92VDD102 pKa = 4.11ASRR105 pKa = 11.84SS106 pKa = 3.56
MM1 pKa = 7.59GIDD4 pKa = 4.45PNFDD8 pKa = 3.82DD9 pKa = 4.06NRR11 pKa = 11.84EE12 pKa = 4.18HH13 pKa = 8.17VGEE16 pKa = 4.12EE17 pKa = 3.65NGLDD21 pKa = 3.13VWGPVDD27 pKa = 3.89PPEE30 pKa = 4.39KK31 pKa = 10.5LGIHH35 pKa = 6.29GAHH38 pKa = 6.12VAVDD42 pKa = 3.77YY43 pKa = 10.01DD44 pKa = 3.37ICIADD49 pKa = 4.36GACLEE54 pKa = 4.2NCPVDD59 pKa = 3.64VFDD62 pKa = 5.23WVDD65 pKa = 3.63TPDD68 pKa = 3.53HH69 pKa = 6.97PEE71 pKa = 3.86SEE73 pKa = 4.48KK74 pKa = 10.81KK75 pKa = 10.45VEE77 pKa = 4.04PTRR80 pKa = 11.84EE81 pKa = 4.05DD82 pKa = 3.22QCIDD86 pKa = 3.44CMLCVDD92 pKa = 4.88ICPVDD97 pKa = 5.13AIDD100 pKa = 3.92VDD102 pKa = 4.11ASRR105 pKa = 11.84SS106 pKa = 3.56
Molecular weight: 11.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5U3J6|A0A1M5U3J6_9EURY Sugar phosphate isomerase/epimerase OS=Halobaculum gomorrense OX=43928 GN=SAMN05443636_2907 PE=4 SV=1
MM1 pKa = 7.78DD2 pKa = 4.19WYY4 pKa = 11.03RR5 pKa = 11.84LLRR8 pKa = 11.84SAGTRR13 pKa = 11.84RR14 pKa = 11.84LSVDD18 pKa = 2.86QRR20 pKa = 11.84ALVALRR26 pKa = 11.84VAVGLLLLTDD36 pKa = 5.03LLLCTRR42 pKa = 11.84SLVFFYY48 pKa = 10.57TDD50 pKa = 2.98GGSLPRR56 pKa = 11.84SALFARR62 pKa = 11.84FPITANLSVHH72 pKa = 6.39ALFGGAWWVGLLFVAAAVAATALALGRR99 pKa = 11.84WTKK102 pKa = 10.34IAAACSLVLLVSLHH116 pKa = 5.79ARR118 pKa = 11.84NPIALNGGDD127 pKa = 3.43SLLRR131 pKa = 11.84RR132 pKa = 11.84TLLWSLFLPLGTGLTFEE149 pKa = 4.56TRR151 pKa = 11.84AQCRR155 pKa = 11.84HH156 pKa = 5.78PAAVGLLVQPIVLYY170 pKa = 10.34VVNAVIKK177 pKa = 10.74LRR179 pKa = 11.84GDD181 pKa = 2.92VWLSGEE187 pKa = 3.79AVKK190 pKa = 10.94YY191 pKa = 8.9VFRR194 pKa = 11.84IDD196 pKa = 3.08ALTRR200 pKa = 11.84FVGEE204 pKa = 3.96YY205 pKa = 9.87LAAYY209 pKa = 6.81PTLLDD214 pKa = 3.82ALGTAWVMLLCCSPLLVLTTGRR236 pKa = 11.84ARR238 pKa = 11.84ATVVAVFASAHH249 pKa = 5.35VGMALTLGIGLFPLIGVAALIPYY272 pKa = 8.51LPPTAWDD279 pKa = 3.44KK280 pKa = 11.98VEE282 pKa = 4.5GSWNRR287 pKa = 11.84SVKK290 pKa = 7.2TARR293 pKa = 11.84RR294 pKa = 11.84VGGEE298 pKa = 3.36RR299 pKa = 11.84RR300 pKa = 11.84AAFEE304 pKa = 4.37DD305 pKa = 3.59LWEE308 pKa = 4.41KK309 pKa = 10.53VCRR312 pKa = 11.84QIGRR316 pKa = 11.84LGRR319 pKa = 11.84GRR321 pKa = 11.84SLGAAPPMKK330 pKa = 10.12PMEE333 pKa = 4.75PLRR336 pKa = 11.84QRR338 pKa = 11.84IGTAGSGGRR347 pKa = 11.84TGSTSSLVSVDD358 pKa = 3.07RR359 pKa = 11.84GRR361 pKa = 11.84IATRR365 pKa = 11.84IRR367 pKa = 11.84RR368 pKa = 11.84IGRR371 pKa = 11.84AVAATMVVVVLIWNTASLGYY391 pKa = 10.4VEE393 pKa = 5.63VPQPEE398 pKa = 4.16AVEE401 pKa = 4.18VSPQEE406 pKa = 4.18TRR408 pKa = 11.84WDD410 pKa = 3.59MFAPSPPTRR419 pKa = 11.84DD420 pKa = 2.59VWYY423 pKa = 10.04VAVGTLEE430 pKa = 4.26DD431 pKa = 3.96GEE433 pKa = 4.34RR434 pKa = 11.84VEE436 pKa = 4.64VIRR439 pKa = 11.84GGEE442 pKa = 4.08PTWARR447 pKa = 11.84SEE449 pKa = 4.42RR450 pKa = 11.84RR451 pKa = 11.84WSSYY455 pKa = 10.36PSARR459 pKa = 11.84WRR461 pKa = 11.84KK462 pKa = 7.98YY463 pKa = 10.08LEE465 pKa = 3.95VVRR468 pKa = 11.84WSDD471 pKa = 3.77DD472 pKa = 3.17DD473 pKa = 3.37WLQRR477 pKa = 11.84HH478 pKa = 5.79FASGLCARR486 pKa = 11.84WDD488 pKa = 3.38ATHH491 pKa = 7.45RR492 pKa = 11.84SRR494 pKa = 11.84LSRR497 pKa = 11.84VTVYY501 pKa = 10.6VLSRR505 pKa = 11.84SVQSSTEE512 pKa = 4.01PNASEE517 pKa = 4.54GPSVRR522 pKa = 11.84RR523 pKa = 11.84TRR525 pKa = 11.84VRR527 pKa = 11.84THH529 pKa = 6.05EE530 pKa = 4.36CSSS533 pKa = 3.28
MM1 pKa = 7.78DD2 pKa = 4.19WYY4 pKa = 11.03RR5 pKa = 11.84LLRR8 pKa = 11.84SAGTRR13 pKa = 11.84RR14 pKa = 11.84LSVDD18 pKa = 2.86QRR20 pKa = 11.84ALVALRR26 pKa = 11.84VAVGLLLLTDD36 pKa = 5.03LLLCTRR42 pKa = 11.84SLVFFYY48 pKa = 10.57TDD50 pKa = 2.98GGSLPRR56 pKa = 11.84SALFARR62 pKa = 11.84FPITANLSVHH72 pKa = 6.39ALFGGAWWVGLLFVAAAVAATALALGRR99 pKa = 11.84WTKK102 pKa = 10.34IAAACSLVLLVSLHH116 pKa = 5.79ARR118 pKa = 11.84NPIALNGGDD127 pKa = 3.43SLLRR131 pKa = 11.84RR132 pKa = 11.84TLLWSLFLPLGTGLTFEE149 pKa = 4.56TRR151 pKa = 11.84AQCRR155 pKa = 11.84HH156 pKa = 5.78PAAVGLLVQPIVLYY170 pKa = 10.34VVNAVIKK177 pKa = 10.74LRR179 pKa = 11.84GDD181 pKa = 2.92VWLSGEE187 pKa = 3.79AVKK190 pKa = 10.94YY191 pKa = 8.9VFRR194 pKa = 11.84IDD196 pKa = 3.08ALTRR200 pKa = 11.84FVGEE204 pKa = 3.96YY205 pKa = 9.87LAAYY209 pKa = 6.81PTLLDD214 pKa = 3.82ALGTAWVMLLCCSPLLVLTTGRR236 pKa = 11.84ARR238 pKa = 11.84ATVVAVFASAHH249 pKa = 5.35VGMALTLGIGLFPLIGVAALIPYY272 pKa = 8.51LPPTAWDD279 pKa = 3.44KK280 pKa = 11.98VEE282 pKa = 4.5GSWNRR287 pKa = 11.84SVKK290 pKa = 7.2TARR293 pKa = 11.84RR294 pKa = 11.84VGGEE298 pKa = 3.36RR299 pKa = 11.84RR300 pKa = 11.84AAFEE304 pKa = 4.37DD305 pKa = 3.59LWEE308 pKa = 4.41KK309 pKa = 10.53VCRR312 pKa = 11.84QIGRR316 pKa = 11.84LGRR319 pKa = 11.84GRR321 pKa = 11.84SLGAAPPMKK330 pKa = 10.12PMEE333 pKa = 4.75PLRR336 pKa = 11.84QRR338 pKa = 11.84IGTAGSGGRR347 pKa = 11.84TGSTSSLVSVDD358 pKa = 3.07RR359 pKa = 11.84GRR361 pKa = 11.84IATRR365 pKa = 11.84IRR367 pKa = 11.84RR368 pKa = 11.84IGRR371 pKa = 11.84AVAATMVVVVLIWNTASLGYY391 pKa = 10.4VEE393 pKa = 5.63VPQPEE398 pKa = 4.16AVEE401 pKa = 4.18VSPQEE406 pKa = 4.18TRR408 pKa = 11.84WDD410 pKa = 3.59MFAPSPPTRR419 pKa = 11.84DD420 pKa = 2.59VWYY423 pKa = 10.04VAVGTLEE430 pKa = 4.26DD431 pKa = 3.96GEE433 pKa = 4.34RR434 pKa = 11.84VEE436 pKa = 4.64VIRR439 pKa = 11.84GGEE442 pKa = 4.08PTWARR447 pKa = 11.84SEE449 pKa = 4.42RR450 pKa = 11.84RR451 pKa = 11.84WSSYY455 pKa = 10.36PSARR459 pKa = 11.84WRR461 pKa = 11.84KK462 pKa = 7.98YY463 pKa = 10.08LEE465 pKa = 3.95VVRR468 pKa = 11.84WSDD471 pKa = 3.77DD472 pKa = 3.17DD473 pKa = 3.37WLQRR477 pKa = 11.84HH478 pKa = 5.79FASGLCARR486 pKa = 11.84WDD488 pKa = 3.38ATHH491 pKa = 7.45RR492 pKa = 11.84SRR494 pKa = 11.84LSRR497 pKa = 11.84VTVYY501 pKa = 10.6VLSRR505 pKa = 11.84SVQSSTEE512 pKa = 4.01PNASEE517 pKa = 4.54GPSVRR522 pKa = 11.84RR523 pKa = 11.84TRR525 pKa = 11.84VRR527 pKa = 11.84THH529 pKa = 6.05EE530 pKa = 4.36CSSS533 pKa = 3.28
Molecular weight: 58.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
922802 |
28 |
2162 |
293.0 |
31.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.492 ± 0.081 | 0.698 ± 0.014 |
8.802 ± 0.057 | 8.083 ± 0.069 |
3.175 ± 0.029 | 9.238 ± 0.042 |
1.937 ± 0.02 | 3.401 ± 0.036 |
1.637 ± 0.027 | 8.672 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.658 ± 0.02 | 2.089 ± 0.026 |
4.778 ± 0.027 | 2.049 ± 0.028 |
6.941 ± 0.05 | 5.231 ± 0.03 |
6.177 ± 0.035 | 9.35 ± 0.047 |
1.086 ± 0.017 | 2.506 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
