
Amphibola crenata associated bacilladnavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; Protobacilladnavirus; Snail associated protobacilladnavirus 2
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
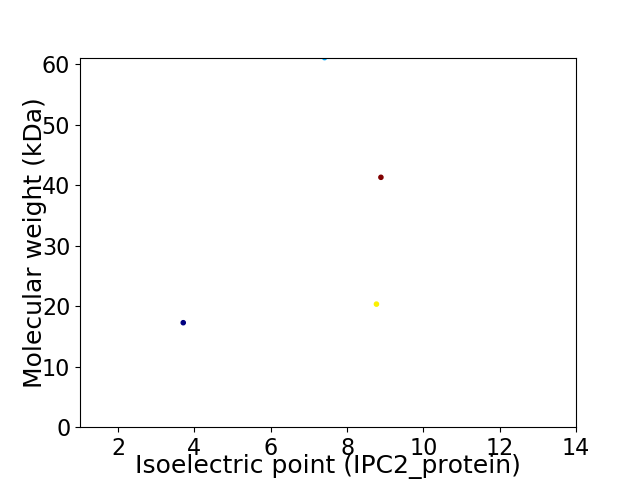
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1P8YT78|A0A1P8YT78_9VIRU p1 OS=Amphibola crenata associated bacilladnavirus 2 OX=1941436 PE=4 SV=1
MM1 pKa = 7.17NMCRR5 pKa = 11.84PILHH9 pKa = 6.43LTAHH13 pKa = 6.06NGVRR17 pKa = 11.84EE18 pKa = 3.88IDD20 pKa = 3.5LSKK23 pKa = 11.11SLNALAMLNMTGFYY37 pKa = 10.87NDD39 pKa = 3.21ADD41 pKa = 4.03YY42 pKa = 11.69YY43 pKa = 11.05GIKK46 pKa = 10.17LWIDD50 pKa = 3.36AYY52 pKa = 10.05MQLANAPMPSSVTFKK67 pKa = 10.54SPCRR71 pKa = 11.84IEE73 pKa = 3.89PVNFITLCSDD83 pKa = 5.06LADD86 pKa = 3.06EE87 pKa = 4.22WAINEE92 pKa = 4.34GQQEE96 pKa = 4.33LQGEE100 pKa = 4.63DD101 pKa = 3.23PAIWADD107 pKa = 3.72EE108 pKa = 4.12VLSISDD114 pKa = 3.54FTVSIDD120 pKa = 4.02DD121 pKa = 4.47LGTIALEE128 pKa = 4.3DD129 pKa = 3.72VNLDD133 pKa = 3.63SFFSNDD139 pKa = 3.07VEE141 pKa = 4.52IMDD144 pKa = 4.7DD145 pKa = 3.47SSVISEE151 pKa = 3.93IVEE154 pKa = 3.88II155 pKa = 4.6
MM1 pKa = 7.17NMCRR5 pKa = 11.84PILHH9 pKa = 6.43LTAHH13 pKa = 6.06NGVRR17 pKa = 11.84EE18 pKa = 3.88IDD20 pKa = 3.5LSKK23 pKa = 11.11SLNALAMLNMTGFYY37 pKa = 10.87NDD39 pKa = 3.21ADD41 pKa = 4.03YY42 pKa = 11.69YY43 pKa = 11.05GIKK46 pKa = 10.17LWIDD50 pKa = 3.36AYY52 pKa = 10.05MQLANAPMPSSVTFKK67 pKa = 10.54SPCRR71 pKa = 11.84IEE73 pKa = 3.89PVNFITLCSDD83 pKa = 5.06LADD86 pKa = 3.06EE87 pKa = 4.22WAINEE92 pKa = 4.34GQQEE96 pKa = 4.33LQGEE100 pKa = 4.63DD101 pKa = 3.23PAIWADD107 pKa = 3.72EE108 pKa = 4.12VLSISDD114 pKa = 3.54FTVSIDD120 pKa = 4.02DD121 pKa = 4.47LGTIALEE128 pKa = 4.3DD129 pKa = 3.72VNLDD133 pKa = 3.63SFFSNDD139 pKa = 3.07VEE141 pKa = 4.52IMDD144 pKa = 4.7DD145 pKa = 3.47SSVISEE151 pKa = 3.93IVEE154 pKa = 3.88II155 pKa = 4.6
Molecular weight: 17.27 kDa
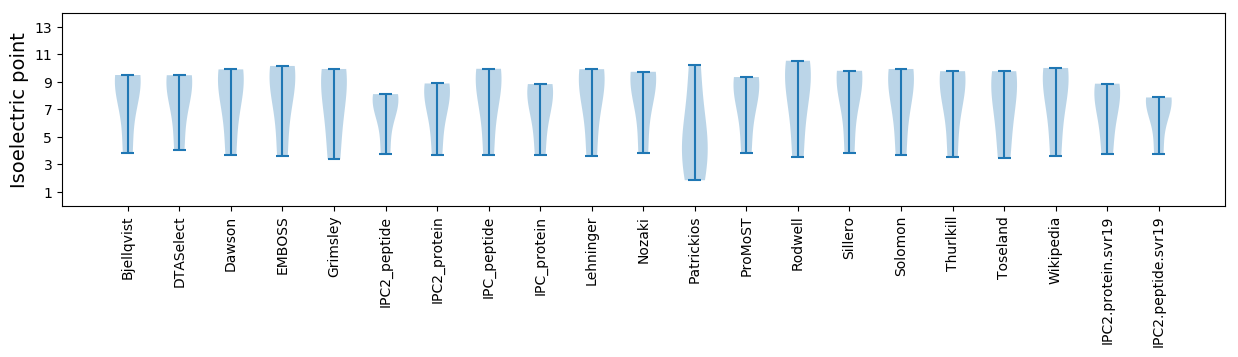
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YT80|A0A1P8YT80_9VIRU Replication-associated protein OS=Amphibola crenata associated bacilladnavirus 2 OX=1941436 PE=4 SV=1
MM1 pKa = 7.61APKK4 pKa = 8.89KK5 pKa = 8.88TPSKK9 pKa = 9.62GRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84TTSVQRR20 pKa = 11.84RR21 pKa = 11.84PNPTKK26 pKa = 10.6RR27 pKa = 11.84KK28 pKa = 5.84PTKK31 pKa = 8.32KK32 pKa = 8.67TKK34 pKa = 9.95KK35 pKa = 10.3AGIAGAAAHH44 pKa = 6.46LLKK47 pKa = 10.37PLLMPFDD54 pKa = 4.17RR55 pKa = 11.84TNMQPKK61 pKa = 9.95IPDD64 pKa = 3.68GKK66 pKa = 10.22VGTSIGQKK74 pKa = 8.47FTTTRR79 pKa = 11.84EE80 pKa = 4.12HH81 pKa = 6.59FNVGAGNIMHH91 pKa = 7.5ALLFPGQSGGLVVIGCQNFNSVPISSPAFNDD122 pKa = 3.11AGGFSYY128 pKa = 8.92THH130 pKa = 7.21DD131 pKa = 4.28LASGMSTLAAKK142 pKa = 10.09EE143 pKa = 4.61NYY145 pKa = 9.44HH146 pKa = 4.99SWRR149 pKa = 11.84VVSQAARR156 pKa = 11.84FEE158 pKa = 4.39LLNAAEE164 pKa = 5.19EE165 pKa = 4.36DD166 pKa = 4.08DD167 pKa = 4.32GWWEE171 pKa = 3.93AVRR174 pKa = 11.84VTDD177 pKa = 4.25SMSVFDD183 pKa = 5.96FRR185 pKa = 11.84LDD187 pKa = 3.32QHH189 pKa = 7.34DD190 pKa = 4.04NGTGRR195 pKa = 11.84GNGTILPYY203 pKa = 9.47YY204 pKa = 9.61TLANYY209 pKa = 7.7QFKK212 pKa = 10.54PITDD216 pKa = 3.21EE217 pKa = 3.75MTYY220 pKa = 10.87EE221 pKa = 4.04SGRR224 pKa = 11.84LKK226 pKa = 10.56DD227 pKa = 3.2LHH229 pKa = 7.34KK230 pKa = 10.87RR231 pKa = 11.84EE232 pKa = 4.21FQLHH236 pKa = 4.06QVKK239 pKa = 11.0DD240 pKa = 3.92EE241 pKa = 4.09VDD243 pKa = 4.03FTQLCTPVTIEE254 pKa = 4.28GGVEE258 pKa = 3.77SGIGIVGSPYY268 pKa = 9.84VVDD271 pKa = 3.58WNGTGGANNGEE282 pKa = 4.21RR283 pKa = 11.84YY284 pKa = 10.31GMQNYY289 pKa = 9.14NAIRR293 pKa = 11.84NYY295 pKa = 10.04IDD297 pKa = 2.87TGYY300 pKa = 11.45DD301 pKa = 2.94MIYY304 pKa = 10.76FRR306 pKa = 11.84FHH308 pKa = 6.3CRR310 pKa = 11.84AAATPTRR317 pKa = 11.84LLTHH321 pKa = 6.45IVSNQEE327 pKa = 3.54IQFSSDD333 pKa = 2.72AGEE336 pKa = 4.38SRR338 pKa = 11.84FQTLSARR345 pKa = 11.84VPQAAATLAARR356 pKa = 11.84ANGGKK361 pKa = 7.52TASVPAQGKK370 pKa = 9.79DD371 pKa = 3.35PSRR374 pKa = 11.84HH375 pKa = 5.34GGG377 pKa = 3.25
MM1 pKa = 7.61APKK4 pKa = 8.89KK5 pKa = 8.88TPSKK9 pKa = 9.62GRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84TTSVQRR20 pKa = 11.84RR21 pKa = 11.84PNPTKK26 pKa = 10.6RR27 pKa = 11.84KK28 pKa = 5.84PTKK31 pKa = 8.32KK32 pKa = 8.67TKK34 pKa = 9.95KK35 pKa = 10.3AGIAGAAAHH44 pKa = 6.46LLKK47 pKa = 10.37PLLMPFDD54 pKa = 4.17RR55 pKa = 11.84TNMQPKK61 pKa = 9.95IPDD64 pKa = 3.68GKK66 pKa = 10.22VGTSIGQKK74 pKa = 8.47FTTTRR79 pKa = 11.84EE80 pKa = 4.12HH81 pKa = 6.59FNVGAGNIMHH91 pKa = 7.5ALLFPGQSGGLVVIGCQNFNSVPISSPAFNDD122 pKa = 3.11AGGFSYY128 pKa = 8.92THH130 pKa = 7.21DD131 pKa = 4.28LASGMSTLAAKK142 pKa = 10.09EE143 pKa = 4.61NYY145 pKa = 9.44HH146 pKa = 4.99SWRR149 pKa = 11.84VVSQAARR156 pKa = 11.84FEE158 pKa = 4.39LLNAAEE164 pKa = 5.19EE165 pKa = 4.36DD166 pKa = 4.08DD167 pKa = 4.32GWWEE171 pKa = 3.93AVRR174 pKa = 11.84VTDD177 pKa = 4.25SMSVFDD183 pKa = 5.96FRR185 pKa = 11.84LDD187 pKa = 3.32QHH189 pKa = 7.34DD190 pKa = 4.04NGTGRR195 pKa = 11.84GNGTILPYY203 pKa = 9.47YY204 pKa = 9.61TLANYY209 pKa = 7.7QFKK212 pKa = 10.54PITDD216 pKa = 3.21EE217 pKa = 3.75MTYY220 pKa = 10.87EE221 pKa = 4.04SGRR224 pKa = 11.84LKK226 pKa = 10.56DD227 pKa = 3.2LHH229 pKa = 7.34KK230 pKa = 10.87RR231 pKa = 11.84EE232 pKa = 4.21FQLHH236 pKa = 4.06QVKK239 pKa = 11.0DD240 pKa = 3.92EE241 pKa = 4.09VDD243 pKa = 4.03FTQLCTPVTIEE254 pKa = 4.28GGVEE258 pKa = 3.77SGIGIVGSPYY268 pKa = 9.84VVDD271 pKa = 3.58WNGTGGANNGEE282 pKa = 4.21RR283 pKa = 11.84YY284 pKa = 10.31GMQNYY289 pKa = 9.14NAIRR293 pKa = 11.84NYY295 pKa = 10.04IDD297 pKa = 2.87TGYY300 pKa = 11.45DD301 pKa = 2.94MIYY304 pKa = 10.76FRR306 pKa = 11.84FHH308 pKa = 6.3CRR310 pKa = 11.84AAATPTRR317 pKa = 11.84LLTHH321 pKa = 6.45IVSNQEE327 pKa = 3.54IQFSSDD333 pKa = 2.72AGEE336 pKa = 4.38SRR338 pKa = 11.84FQTLSARR345 pKa = 11.84VPQAAATLAARR356 pKa = 11.84ANGGKK361 pKa = 7.52TASVPAQGKK370 pKa = 9.79DD371 pKa = 3.35PSRR374 pKa = 11.84HH375 pKa = 5.34GGG377 pKa = 3.25
Molecular weight: 41.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1242 |
155 |
532 |
310.5 |
35.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.683 ± 1.189 | 1.369 ± 0.393 |
6.441 ± 0.963 | 6.441 ± 0.898 |
4.267 ± 0.455 | 7.246 ± 1.115 |
2.979 ± 0.548 | 6.039 ± 0.906 |
7.166 ± 1.796 | 6.039 ± 0.841 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.392 | 4.348 ± 0.726 |
4.992 ± 0.199 | 3.301 ± 0.469 |
5.878 ± 0.745 | 7.085 ± 0.454 |
7.005 ± 0.597 | 5.314 ± 0.165 |
1.771 ± 0.437 | 3.14 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
