
Beet cryptic virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
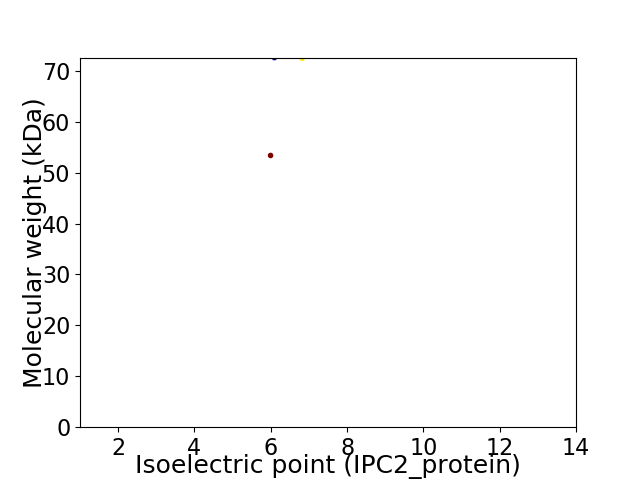
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2CGP4|B2CGP4_9VIRU Coat protein OS=Beet cryptic virus 1 OX=509923 GN=CP PE=4 SV=1
MM1 pKa = 7.65EE2 pKa = 5.7NNTPLANPSGPNVPSAAAPPTPAPPAIPQATPTPGAVSQPPAPPARR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84TPRR54 pKa = 11.84GPVPQATPGSTSGAPALLEE73 pKa = 4.05LSAGLPMYY81 pKa = 7.42TVPRR85 pKa = 11.84RR86 pKa = 11.84GVNTFVPDD94 pKa = 3.46AQMLFHH100 pKa = 6.86VLGICDD106 pKa = 3.86QMMLSTDD113 pKa = 3.09RR114 pKa = 11.84FTRR117 pKa = 11.84SSPAWIPIVSQLYY130 pKa = 10.22ISVLWMVAILRR141 pKa = 11.84VFVASGYY148 pKa = 9.09GALYY152 pKa = 10.6SSLINDD158 pKa = 5.42LIGHH162 pKa = 6.3LRR164 pKa = 11.84IDD166 pKa = 3.56EE167 pKa = 4.15CMIPGPLVPFFQSLGAVCGPYY188 pKa = 10.28EE189 pKa = 4.28WIGDD193 pKa = 3.7IVAAFPDD200 pKa = 5.36FLTLWDD206 pKa = 4.75AEE208 pKa = 4.59NFCPTADD215 pKa = 3.98LARR218 pKa = 11.84TCPVPAIMLDD228 pKa = 3.39QLHH231 pKa = 6.39YY232 pKa = 10.03FATWTIPAEE241 pKa = 4.1QILYY245 pKa = 10.04TNFQWYY251 pKa = 10.28RR252 pKa = 11.84NIFSLGLGAGNANNRR267 pKa = 11.84IGPQLCGSLYY277 pKa = 10.52SPRR280 pKa = 11.84AQVDD284 pKa = 3.47SARR287 pKa = 11.84AFWNAALSSGITRR300 pKa = 11.84TNAAEE305 pKa = 4.07ANGAFYY311 pKa = 9.67TYY313 pKa = 10.96AQLLGFISQNGTLQLDD329 pKa = 4.28WFQQVAVVMQKK340 pKa = 8.58YY341 pKa = 6.0TQYY344 pKa = 11.03FNGSTPLKK352 pKa = 9.96SISTIGIGAVAVIGAPTPDD371 pKa = 3.91PATRR375 pKa = 11.84DD376 pKa = 3.13WFYY379 pKa = 10.7PAATGIEE386 pKa = 4.16PFLCSRR392 pKa = 11.84FAPRR396 pKa = 11.84RR397 pKa = 11.84EE398 pKa = 3.99IPNTLGMIFSHH409 pKa = 7.11ADD411 pKa = 3.1HH412 pKa = 6.65EE413 pKa = 4.56LEE415 pKa = 4.25EE416 pKa = 4.19QAEE419 pKa = 4.33QYY421 pKa = 11.25AILTHH426 pKa = 6.0TNIRR430 pKa = 11.84WSPSVVAQNAWTAVNDD446 pKa = 4.24GASRR450 pKa = 11.84NGDD453 pKa = 3.25YY454 pKa = 10.44WIMMNYY460 pKa = 9.8RR461 pKa = 11.84FSTRR465 pKa = 11.84ISLKK469 pKa = 7.94TQFAQVIASRR479 pKa = 11.84YY480 pKa = 7.5HH481 pKa = 4.12QQAANRR487 pKa = 11.84VDD489 pKa = 3.2
MM1 pKa = 7.65EE2 pKa = 5.7NNTPLANPSGPNVPSAAAPPTPAPPAIPQATPTPGAVSQPPAPPARR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84TPRR54 pKa = 11.84GPVPQATPGSTSGAPALLEE73 pKa = 4.05LSAGLPMYY81 pKa = 7.42TVPRR85 pKa = 11.84RR86 pKa = 11.84GVNTFVPDD94 pKa = 3.46AQMLFHH100 pKa = 6.86VLGICDD106 pKa = 3.86QMMLSTDD113 pKa = 3.09RR114 pKa = 11.84FTRR117 pKa = 11.84SSPAWIPIVSQLYY130 pKa = 10.22ISVLWMVAILRR141 pKa = 11.84VFVASGYY148 pKa = 9.09GALYY152 pKa = 10.6SSLINDD158 pKa = 5.42LIGHH162 pKa = 6.3LRR164 pKa = 11.84IDD166 pKa = 3.56EE167 pKa = 4.15CMIPGPLVPFFQSLGAVCGPYY188 pKa = 10.28EE189 pKa = 4.28WIGDD193 pKa = 3.7IVAAFPDD200 pKa = 5.36FLTLWDD206 pKa = 4.75AEE208 pKa = 4.59NFCPTADD215 pKa = 3.98LARR218 pKa = 11.84TCPVPAIMLDD228 pKa = 3.39QLHH231 pKa = 6.39YY232 pKa = 10.03FATWTIPAEE241 pKa = 4.1QILYY245 pKa = 10.04TNFQWYY251 pKa = 10.28RR252 pKa = 11.84NIFSLGLGAGNANNRR267 pKa = 11.84IGPQLCGSLYY277 pKa = 10.52SPRR280 pKa = 11.84AQVDD284 pKa = 3.47SARR287 pKa = 11.84AFWNAALSSGITRR300 pKa = 11.84TNAAEE305 pKa = 4.07ANGAFYY311 pKa = 9.67TYY313 pKa = 10.96AQLLGFISQNGTLQLDD329 pKa = 4.28WFQQVAVVMQKK340 pKa = 8.58YY341 pKa = 6.0TQYY344 pKa = 11.03FNGSTPLKK352 pKa = 9.96SISTIGIGAVAVIGAPTPDD371 pKa = 3.91PATRR375 pKa = 11.84DD376 pKa = 3.13WFYY379 pKa = 10.7PAATGIEE386 pKa = 4.16PFLCSRR392 pKa = 11.84FAPRR396 pKa = 11.84RR397 pKa = 11.84EE398 pKa = 3.99IPNTLGMIFSHH409 pKa = 7.11ADD411 pKa = 3.1HH412 pKa = 6.65EE413 pKa = 4.56LEE415 pKa = 4.25EE416 pKa = 4.19QAEE419 pKa = 4.33QYY421 pKa = 11.25AILTHH426 pKa = 6.0TNIRR430 pKa = 11.84WSPSVVAQNAWTAVNDD446 pKa = 4.24GASRR450 pKa = 11.84NGDD453 pKa = 3.25YY454 pKa = 10.44WIMMNYY460 pKa = 9.8RR461 pKa = 11.84FSTRR465 pKa = 11.84ISLKK469 pKa = 7.94TQFAQVIASRR479 pKa = 11.84YY480 pKa = 7.5HH481 pKa = 4.12QQAANRR487 pKa = 11.84VDD489 pKa = 3.2
Molecular weight: 53.36 kDa
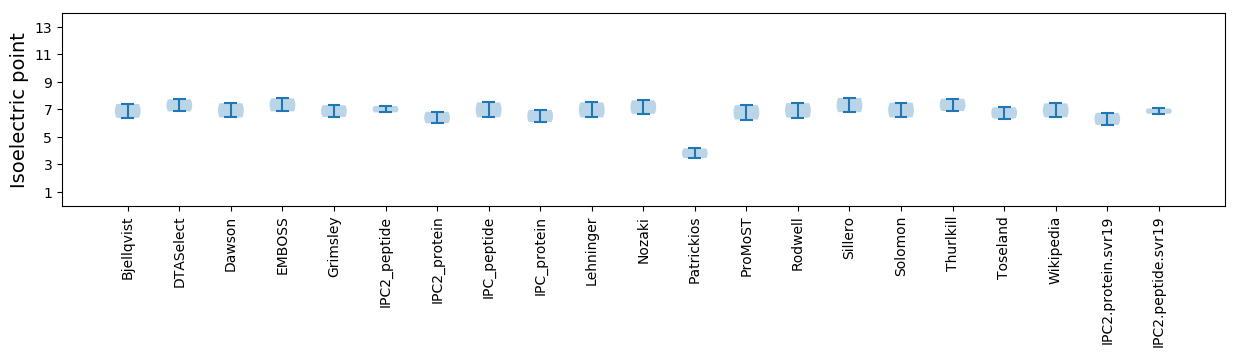
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2CGP4|B2CGP4_9VIRU Coat protein OS=Beet cryptic virus 1 OX=509923 GN=CP PE=4 SV=1
MM1 pKa = 8.32DD2 pKa = 4.54YY3 pKa = 9.87LTSAFNRR10 pKa = 11.84ITHH13 pKa = 6.46WFNVPSNLEE22 pKa = 4.12YY23 pKa = 10.43IGTFSLPPGILRR35 pKa = 11.84VNEE38 pKa = 3.98VAISNHH44 pKa = 5.66KK45 pKa = 8.79KK46 pKa = 8.16TLEE49 pKa = 3.89HH50 pKa = 6.7SFNKK54 pKa = 9.98YY55 pKa = 10.23LYY57 pKa = 8.7AHH59 pKa = 7.41EE60 pKa = 5.06IKK62 pKa = 10.74LITQDD67 pKa = 3.55YY68 pKa = 9.07RR69 pKa = 11.84RR70 pKa = 11.84SDD72 pKa = 3.13IDD74 pKa = 3.64EE75 pKa = 4.23EE76 pKa = 4.91SILADD81 pKa = 4.29FFSGDD86 pKa = 3.36VEE88 pKa = 4.33KK89 pKa = 11.31FEE91 pKa = 4.84VPFDD95 pKa = 3.62EE96 pKa = 5.13HH97 pKa = 8.54VEE99 pKa = 4.09TGLRR103 pKa = 11.84CMADD107 pKa = 2.92AFRR110 pKa = 11.84PPRR113 pKa = 11.84LCRR116 pKa = 11.84PAHH119 pKa = 6.03ILDD122 pKa = 3.78VKK124 pKa = 10.61HH125 pKa = 6.44GYY127 pKa = 8.11PYY129 pKa = 10.43KK130 pKa = 10.06WNVNAEE136 pKa = 4.36PPFSTDD142 pKa = 3.36EE143 pKa = 4.02YY144 pKa = 11.11FLNQRR149 pKa = 11.84KK150 pKa = 7.03TFGEE154 pKa = 4.96FIRR157 pKa = 11.84MHH159 pKa = 6.57EE160 pKa = 4.18YY161 pKa = 10.71EE162 pKa = 5.8HH163 pKa = 7.06IDD165 pKa = 3.09KK166 pKa = 11.16ADD168 pKa = 3.68FFRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 6.43PNTEE177 pKa = 3.6SHH179 pKa = 6.91DD180 pKa = 4.58LIRR183 pKa = 11.84TIVPPKK189 pKa = 10.24FGYY192 pKa = 9.49LKK194 pKa = 11.12SMIFSWTRR202 pKa = 11.84RR203 pKa = 11.84WHH205 pKa = 6.41HH206 pKa = 6.48IIKK209 pKa = 10.51EE210 pKa = 4.24GFTEE214 pKa = 4.22STGLHH219 pKa = 4.48TTGYY223 pKa = 9.46FYY225 pKa = 11.38NRR227 pKa = 11.84FIFPMLLHH235 pKa = 6.66TKK237 pKa = 8.28TAIVKK242 pKa = 10.05QDD244 pKa = 4.31DD245 pKa = 4.02PNKK248 pKa = 9.92MRR250 pKa = 11.84TIWGASKK257 pKa = 9.78PWIIAEE263 pKa = 4.12TMLYY267 pKa = 9.2WEE269 pKa = 4.49YY270 pKa = 10.8IAWIKK275 pKa = 10.47LNPSVTPMLWGYY287 pKa = 7.52EE288 pKa = 4.17TFTGGWFRR296 pKa = 11.84LNRR299 pKa = 11.84DD300 pKa = 4.2LFCGFLQRR308 pKa = 11.84SFLTLDD314 pKa = 3.43WSRR317 pKa = 11.84FDD319 pKa = 3.15KK320 pKa = 10.78RR321 pKa = 11.84AYY323 pKa = 10.14FPLLRR328 pKa = 11.84RR329 pKa = 11.84ILYY332 pKa = 6.53TARR335 pKa = 11.84TFLTFDD341 pKa = 2.91EE342 pKa = 6.12GYY344 pKa = 10.56VPTHH348 pKa = 6.37SAPTHH353 pKa = 5.49PQWDD357 pKa = 3.61HH358 pKa = 5.16TKK360 pKa = 10.6AIRR363 pKa = 11.84LEE365 pKa = 4.25RR366 pKa = 11.84LWLWTLEE373 pKa = 4.18NLFEE377 pKa = 4.94APIILPDD384 pKa = 2.89GRR386 pKa = 11.84MYY388 pKa = 10.65RR389 pKa = 11.84RR390 pKa = 11.84HH391 pKa = 5.78FAGIPSGLFITQLLDD406 pKa = 2.7SWYY409 pKa = 9.85NYY411 pKa = 9.33TMLATLLSALGFDD424 pKa = 4.53PKK426 pKa = 10.86HH427 pKa = 6.68CIIKK431 pKa = 10.05VQGDD435 pKa = 3.54DD436 pKa = 3.82SIIRR440 pKa = 11.84LNVLVPQDD448 pKa = 3.3QHH450 pKa = 7.33QNLMDD455 pKa = 4.24NLVQLAVNYY464 pKa = 9.3FNAVVNVKK472 pKa = 10.2KK473 pKa = 10.96SEE475 pKa = 4.14FGNSLNGRR483 pKa = 11.84EE484 pKa = 3.96VLSYY488 pKa = 10.61RR489 pKa = 11.84NHH491 pKa = 6.46NGFPHH496 pKa = 7.07RR497 pKa = 11.84DD498 pKa = 3.77EE499 pKa = 5.66IMMLAQFYY507 pKa = 7.88HH508 pKa = 6.11TKK510 pKa = 10.61ARR512 pKa = 11.84DD513 pKa = 3.58PTPEE517 pKa = 3.33ITMAQAIGFAYY528 pKa = 10.22ASCANNKK535 pKa = 9.15RR536 pKa = 11.84VLWALKK542 pKa = 10.2DD543 pKa = 3.49VYY545 pKa = 10.88DD546 pKa = 3.87YY547 pKa = 11.81YY548 pKa = 11.61KK549 pKa = 11.07DD550 pKa = 3.95LGYY553 pKa = 9.29TPNRR557 pKa = 11.84AGLTLTFGDD566 pKa = 4.33SPDD569 pKa = 3.58LFVPEE574 pKa = 4.48ISLEE578 pKa = 4.07HH579 pKa = 6.61FPTEE583 pKa = 4.19TEE585 pKa = 3.13IRR587 pKa = 11.84RR588 pKa = 11.84YY589 pKa = 8.3LTSTSYY595 pKa = 11.44LNEE598 pKa = 4.1AQNARR603 pKa = 11.84TWPRR607 pKa = 11.84TLFINAPAQQ616 pKa = 3.31
MM1 pKa = 8.32DD2 pKa = 4.54YY3 pKa = 9.87LTSAFNRR10 pKa = 11.84ITHH13 pKa = 6.46WFNVPSNLEE22 pKa = 4.12YY23 pKa = 10.43IGTFSLPPGILRR35 pKa = 11.84VNEE38 pKa = 3.98VAISNHH44 pKa = 5.66KK45 pKa = 8.79KK46 pKa = 8.16TLEE49 pKa = 3.89HH50 pKa = 6.7SFNKK54 pKa = 9.98YY55 pKa = 10.23LYY57 pKa = 8.7AHH59 pKa = 7.41EE60 pKa = 5.06IKK62 pKa = 10.74LITQDD67 pKa = 3.55YY68 pKa = 9.07RR69 pKa = 11.84RR70 pKa = 11.84SDD72 pKa = 3.13IDD74 pKa = 3.64EE75 pKa = 4.23EE76 pKa = 4.91SILADD81 pKa = 4.29FFSGDD86 pKa = 3.36VEE88 pKa = 4.33KK89 pKa = 11.31FEE91 pKa = 4.84VPFDD95 pKa = 3.62EE96 pKa = 5.13HH97 pKa = 8.54VEE99 pKa = 4.09TGLRR103 pKa = 11.84CMADD107 pKa = 2.92AFRR110 pKa = 11.84PPRR113 pKa = 11.84LCRR116 pKa = 11.84PAHH119 pKa = 6.03ILDD122 pKa = 3.78VKK124 pKa = 10.61HH125 pKa = 6.44GYY127 pKa = 8.11PYY129 pKa = 10.43KK130 pKa = 10.06WNVNAEE136 pKa = 4.36PPFSTDD142 pKa = 3.36EE143 pKa = 4.02YY144 pKa = 11.11FLNQRR149 pKa = 11.84KK150 pKa = 7.03TFGEE154 pKa = 4.96FIRR157 pKa = 11.84MHH159 pKa = 6.57EE160 pKa = 4.18YY161 pKa = 10.71EE162 pKa = 5.8HH163 pKa = 7.06IDD165 pKa = 3.09KK166 pKa = 11.16ADD168 pKa = 3.68FFRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 6.43PNTEE177 pKa = 3.6SHH179 pKa = 6.91DD180 pKa = 4.58LIRR183 pKa = 11.84TIVPPKK189 pKa = 10.24FGYY192 pKa = 9.49LKK194 pKa = 11.12SMIFSWTRR202 pKa = 11.84RR203 pKa = 11.84WHH205 pKa = 6.41HH206 pKa = 6.48IIKK209 pKa = 10.51EE210 pKa = 4.24GFTEE214 pKa = 4.22STGLHH219 pKa = 4.48TTGYY223 pKa = 9.46FYY225 pKa = 11.38NRR227 pKa = 11.84FIFPMLLHH235 pKa = 6.66TKK237 pKa = 8.28TAIVKK242 pKa = 10.05QDD244 pKa = 4.31DD245 pKa = 4.02PNKK248 pKa = 9.92MRR250 pKa = 11.84TIWGASKK257 pKa = 9.78PWIIAEE263 pKa = 4.12TMLYY267 pKa = 9.2WEE269 pKa = 4.49YY270 pKa = 10.8IAWIKK275 pKa = 10.47LNPSVTPMLWGYY287 pKa = 7.52EE288 pKa = 4.17TFTGGWFRR296 pKa = 11.84LNRR299 pKa = 11.84DD300 pKa = 4.2LFCGFLQRR308 pKa = 11.84SFLTLDD314 pKa = 3.43WSRR317 pKa = 11.84FDD319 pKa = 3.15KK320 pKa = 10.78RR321 pKa = 11.84AYY323 pKa = 10.14FPLLRR328 pKa = 11.84RR329 pKa = 11.84ILYY332 pKa = 6.53TARR335 pKa = 11.84TFLTFDD341 pKa = 2.91EE342 pKa = 6.12GYY344 pKa = 10.56VPTHH348 pKa = 6.37SAPTHH353 pKa = 5.49PQWDD357 pKa = 3.61HH358 pKa = 5.16TKK360 pKa = 10.6AIRR363 pKa = 11.84LEE365 pKa = 4.25RR366 pKa = 11.84LWLWTLEE373 pKa = 4.18NLFEE377 pKa = 4.94APIILPDD384 pKa = 2.89GRR386 pKa = 11.84MYY388 pKa = 10.65RR389 pKa = 11.84RR390 pKa = 11.84HH391 pKa = 5.78FAGIPSGLFITQLLDD406 pKa = 2.7SWYY409 pKa = 9.85NYY411 pKa = 9.33TMLATLLSALGFDD424 pKa = 4.53PKK426 pKa = 10.86HH427 pKa = 6.68CIIKK431 pKa = 10.05VQGDD435 pKa = 3.54DD436 pKa = 3.82SIIRR440 pKa = 11.84LNVLVPQDD448 pKa = 3.3QHH450 pKa = 7.33QNLMDD455 pKa = 4.24NLVQLAVNYY464 pKa = 9.3FNAVVNVKK472 pKa = 10.2KK473 pKa = 10.96SEE475 pKa = 4.14FGNSLNGRR483 pKa = 11.84EE484 pKa = 3.96VLSYY488 pKa = 10.61RR489 pKa = 11.84NHH491 pKa = 6.46NGFPHH496 pKa = 7.07RR497 pKa = 11.84DD498 pKa = 3.77EE499 pKa = 5.66IMMLAQFYY507 pKa = 7.88HH508 pKa = 6.11TKK510 pKa = 10.61ARR512 pKa = 11.84DD513 pKa = 3.58PTPEE517 pKa = 3.33ITMAQAIGFAYY528 pKa = 10.22ASCANNKK535 pKa = 9.15RR536 pKa = 11.84VLWALKK542 pKa = 10.2DD543 pKa = 3.49VYY545 pKa = 10.88DD546 pKa = 3.87YY547 pKa = 11.81YY548 pKa = 11.61KK549 pKa = 11.07DD550 pKa = 3.95LGYY553 pKa = 9.29TPNRR557 pKa = 11.84AGLTLTFGDD566 pKa = 4.33SPDD569 pKa = 3.58LFVPEE574 pKa = 4.48ISLEE578 pKa = 4.07HH579 pKa = 6.61FPTEE583 pKa = 4.19TEE585 pKa = 3.13IRR587 pKa = 11.84RR588 pKa = 11.84YY589 pKa = 8.3LTSTSYY595 pKa = 11.44LNEE598 pKa = 4.1AQNARR603 pKa = 11.84TWPRR607 pKa = 11.84TLFINAPAQQ616 pKa = 3.31
Molecular weight: 72.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1105 |
489 |
616 |
552.5 |
62.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.597 ± 2.231 | 1.086 ± 0.222 |
4.706 ± 0.659 | 4.253 ± 1.026 |
5.973 ± 0.817 | 5.43 ± 0.717 |
2.896 ± 0.942 | 6.335 ± 0.003 |
2.715 ± 1.352 | 8.869 ± 0.706 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.353 ± 0.065 | 5.068 ± 0.103 |
7.421 ± 1.278 | 3.71 ± 1.033 |
6.063 ± 0.48 | 5.792 ± 0.747 |
7.059 ± 0.2 | 4.615 ± 0.583 |
2.624 ± 0.11 | 4.434 ± 0.485 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
