
Invertebrate iridescent virus 3 (IIV-3) (Mosquito iridescent virus)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Iridoviridae; Betairidovirinae; Chloriridovirus
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
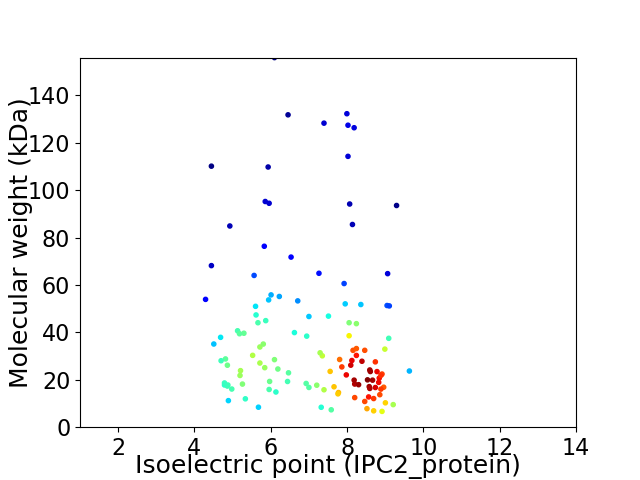
Virtual 2D-PAGE plot for 126 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q197F9|VF395_IIV3 Uncharacterized protein 001R OS=Invertebrate iridescent virus 3 OX=345201 GN=IIV3-001R PE=3 SV=1
MM1 pKa = 7.83ASNTVSAQGGSNRR14 pKa = 11.84PVRR17 pKa = 11.84DD18 pKa = 3.99FSNIQDD24 pKa = 3.47VAQFLLFDD32 pKa = 5.76PIWNEE37 pKa = 3.76QPGSIVPWKK46 pKa = 9.76MNRR49 pKa = 11.84EE50 pKa = 3.52QALAEE55 pKa = 4.57RR56 pKa = 11.84YY57 pKa = 9.34PEE59 pKa = 4.31LQTSEE64 pKa = 4.24PSEE67 pKa = 4.8DD68 pKa = 3.29YY69 pKa = 10.95SGPVEE74 pKa = 4.3SLEE77 pKa = 4.59LLPLEE82 pKa = 4.42IKK84 pKa = 10.52LDD86 pKa = 3.31IMQYY90 pKa = 10.86LSWEE94 pKa = 4.03QISWCKK100 pKa = 10.51HH101 pKa = 3.79PWLWTRR107 pKa = 11.84WYY109 pKa = 9.35KK110 pKa = 11.4DD111 pKa = 2.97NVVRR115 pKa = 11.84VSAITFEE122 pKa = 4.31DD123 pKa = 4.23FQRR126 pKa = 11.84EE127 pKa = 3.88YY128 pKa = 11.45AFPEE132 pKa = 4.63KK133 pKa = 10.04IQEE136 pKa = 3.74IHH138 pKa = 5.8FTDD141 pKa = 3.56TRR143 pKa = 11.84AEE145 pKa = 4.12EE146 pKa = 3.83IKK148 pKa = 10.69AILEE152 pKa = 4.05TTPNVTRR159 pKa = 11.84LVIRR163 pKa = 11.84RR164 pKa = 11.84IDD166 pKa = 3.48DD167 pKa = 3.38MNYY170 pKa = 8.06NTHH173 pKa = 6.75GDD175 pKa = 3.61LGLDD179 pKa = 3.25DD180 pKa = 6.25LEE182 pKa = 4.78FLTHH186 pKa = 7.12LMVEE190 pKa = 4.59DD191 pKa = 3.58ACGFTDD197 pKa = 4.98FWAPSLTHH205 pKa = 6.21LTIKK209 pKa = 10.42NLDD212 pKa = 3.26MHH214 pKa = 6.54PRR216 pKa = 11.84WFGPVMDD223 pKa = 6.2GIKK226 pKa = 10.61SMQSTLKK233 pKa = 10.13YY234 pKa = 10.38LYY236 pKa = 9.88IFEE239 pKa = 4.7TYY241 pKa = 10.04GVNKK245 pKa = 9.92PFVQWCTDD253 pKa = 3.54NIEE256 pKa = 4.33TFYY259 pKa = 10.49CTNSYY264 pKa = 10.33RR265 pKa = 11.84YY266 pKa = 9.28EE267 pKa = 4.06NVPRR271 pKa = 11.84PIYY274 pKa = 10.44VWVLFQEE281 pKa = 4.93DD282 pKa = 3.58EE283 pKa = 3.92WHH285 pKa = 6.76GYY287 pKa = 8.4RR288 pKa = 11.84VEE290 pKa = 4.19DD291 pKa = 3.39NKK293 pKa = 10.19FHH295 pKa = 6.72RR296 pKa = 11.84RR297 pKa = 11.84YY298 pKa = 8.61MYY300 pKa = 9.69STILHH305 pKa = 6.22KK306 pKa = 10.47RR307 pKa = 11.84DD308 pKa = 3.52TDD310 pKa = 3.51WVEE313 pKa = 3.87NNPLKK318 pKa = 10.3TPAQVEE324 pKa = 4.56MYY326 pKa = 10.32KK327 pKa = 10.55FLLRR331 pKa = 11.84ISQLNRR337 pKa = 11.84DD338 pKa = 3.6GTGYY342 pKa = 10.78EE343 pKa = 4.08SDD345 pKa = 3.54SDD347 pKa = 4.1PEE349 pKa = 4.16NEE351 pKa = 4.48HH352 pKa = 6.87FDD354 pKa = 4.21DD355 pKa = 4.66EE356 pKa = 4.89SFSSGEE362 pKa = 3.99EE363 pKa = 4.03DD364 pKa = 5.31SSDD367 pKa = 3.6EE368 pKa = 5.6DD369 pKa = 4.11DD370 pKa = 4.44PTWAPDD376 pKa = 3.51SDD378 pKa = 3.94DD379 pKa = 4.79SDD381 pKa = 3.73WEE383 pKa = 4.49TEE385 pKa = 4.2TEE387 pKa = 4.19EE388 pKa = 4.45EE389 pKa = 4.21PSVAARR395 pKa = 11.84ILEE398 pKa = 4.25KK399 pKa = 10.97GKK401 pKa = 8.91LTITNLMKK409 pKa = 10.84SLGFKK414 pKa = 10.09PKK416 pKa = 9.46PKK418 pKa = 9.99KK419 pKa = 9.46IQSIDD424 pKa = 3.33RR425 pKa = 11.84YY426 pKa = 10.31FCSLDD431 pKa = 3.4SNYY434 pKa = 10.45NSEE437 pKa = 5.38DD438 pKa = 3.25EE439 pKa = 4.32DD440 pKa = 4.12FEE442 pKa = 4.73YY443 pKa = 10.99DD444 pKa = 4.65SDD446 pKa = 5.66SEE448 pKa = 5.57DD449 pKa = 4.43DD450 pKa = 5.17DD451 pKa = 5.62SDD453 pKa = 5.89SEE455 pKa = 4.5DD456 pKa = 3.46DD457 pKa = 3.8CC458 pKa = 7.55
MM1 pKa = 7.83ASNTVSAQGGSNRR14 pKa = 11.84PVRR17 pKa = 11.84DD18 pKa = 3.99FSNIQDD24 pKa = 3.47VAQFLLFDD32 pKa = 5.76PIWNEE37 pKa = 3.76QPGSIVPWKK46 pKa = 9.76MNRR49 pKa = 11.84EE50 pKa = 3.52QALAEE55 pKa = 4.57RR56 pKa = 11.84YY57 pKa = 9.34PEE59 pKa = 4.31LQTSEE64 pKa = 4.24PSEE67 pKa = 4.8DD68 pKa = 3.29YY69 pKa = 10.95SGPVEE74 pKa = 4.3SLEE77 pKa = 4.59LLPLEE82 pKa = 4.42IKK84 pKa = 10.52LDD86 pKa = 3.31IMQYY90 pKa = 10.86LSWEE94 pKa = 4.03QISWCKK100 pKa = 10.51HH101 pKa = 3.79PWLWTRR107 pKa = 11.84WYY109 pKa = 9.35KK110 pKa = 11.4DD111 pKa = 2.97NVVRR115 pKa = 11.84VSAITFEE122 pKa = 4.31DD123 pKa = 4.23FQRR126 pKa = 11.84EE127 pKa = 3.88YY128 pKa = 11.45AFPEE132 pKa = 4.63KK133 pKa = 10.04IQEE136 pKa = 3.74IHH138 pKa = 5.8FTDD141 pKa = 3.56TRR143 pKa = 11.84AEE145 pKa = 4.12EE146 pKa = 3.83IKK148 pKa = 10.69AILEE152 pKa = 4.05TTPNVTRR159 pKa = 11.84LVIRR163 pKa = 11.84RR164 pKa = 11.84IDD166 pKa = 3.48DD167 pKa = 3.38MNYY170 pKa = 8.06NTHH173 pKa = 6.75GDD175 pKa = 3.61LGLDD179 pKa = 3.25DD180 pKa = 6.25LEE182 pKa = 4.78FLTHH186 pKa = 7.12LMVEE190 pKa = 4.59DD191 pKa = 3.58ACGFTDD197 pKa = 4.98FWAPSLTHH205 pKa = 6.21LTIKK209 pKa = 10.42NLDD212 pKa = 3.26MHH214 pKa = 6.54PRR216 pKa = 11.84WFGPVMDD223 pKa = 6.2GIKK226 pKa = 10.61SMQSTLKK233 pKa = 10.13YY234 pKa = 10.38LYY236 pKa = 9.88IFEE239 pKa = 4.7TYY241 pKa = 10.04GVNKK245 pKa = 9.92PFVQWCTDD253 pKa = 3.54NIEE256 pKa = 4.33TFYY259 pKa = 10.49CTNSYY264 pKa = 10.33RR265 pKa = 11.84YY266 pKa = 9.28EE267 pKa = 4.06NVPRR271 pKa = 11.84PIYY274 pKa = 10.44VWVLFQEE281 pKa = 4.93DD282 pKa = 3.58EE283 pKa = 3.92WHH285 pKa = 6.76GYY287 pKa = 8.4RR288 pKa = 11.84VEE290 pKa = 4.19DD291 pKa = 3.39NKK293 pKa = 10.19FHH295 pKa = 6.72RR296 pKa = 11.84RR297 pKa = 11.84YY298 pKa = 8.61MYY300 pKa = 9.69STILHH305 pKa = 6.22KK306 pKa = 10.47RR307 pKa = 11.84DD308 pKa = 3.52TDD310 pKa = 3.51WVEE313 pKa = 3.87NNPLKK318 pKa = 10.3TPAQVEE324 pKa = 4.56MYY326 pKa = 10.32KK327 pKa = 10.55FLLRR331 pKa = 11.84ISQLNRR337 pKa = 11.84DD338 pKa = 3.6GTGYY342 pKa = 10.78EE343 pKa = 4.08SDD345 pKa = 3.54SDD347 pKa = 4.1PEE349 pKa = 4.16NEE351 pKa = 4.48HH352 pKa = 6.87FDD354 pKa = 4.21DD355 pKa = 4.66EE356 pKa = 4.89SFSSGEE362 pKa = 3.99EE363 pKa = 4.03DD364 pKa = 5.31SSDD367 pKa = 3.6EE368 pKa = 5.6DD369 pKa = 4.11DD370 pKa = 4.44PTWAPDD376 pKa = 3.51SDD378 pKa = 3.94DD379 pKa = 4.79SDD381 pKa = 3.73WEE383 pKa = 4.49TEE385 pKa = 4.2TEE387 pKa = 4.19EE388 pKa = 4.45EE389 pKa = 4.21PSVAARR395 pKa = 11.84ILEE398 pKa = 4.25KK399 pKa = 10.97GKK401 pKa = 8.91LTITNLMKK409 pKa = 10.84SLGFKK414 pKa = 10.09PKK416 pKa = 9.46PKK418 pKa = 9.99KK419 pKa = 9.46IQSIDD424 pKa = 3.33RR425 pKa = 11.84YY426 pKa = 10.31FCSLDD431 pKa = 3.4SNYY434 pKa = 10.45NSEE437 pKa = 5.38DD438 pKa = 3.25EE439 pKa = 4.32DD440 pKa = 4.12FEE442 pKa = 4.73YY443 pKa = 10.99DD444 pKa = 4.65SDD446 pKa = 5.66SEE448 pKa = 5.57DD449 pKa = 4.43DD450 pKa = 5.17DD451 pKa = 5.62SDD453 pKa = 5.89SEE455 pKa = 4.5DD456 pKa = 3.46DD457 pKa = 3.8CC458 pKa = 7.55
Molecular weight: 53.92 kDa
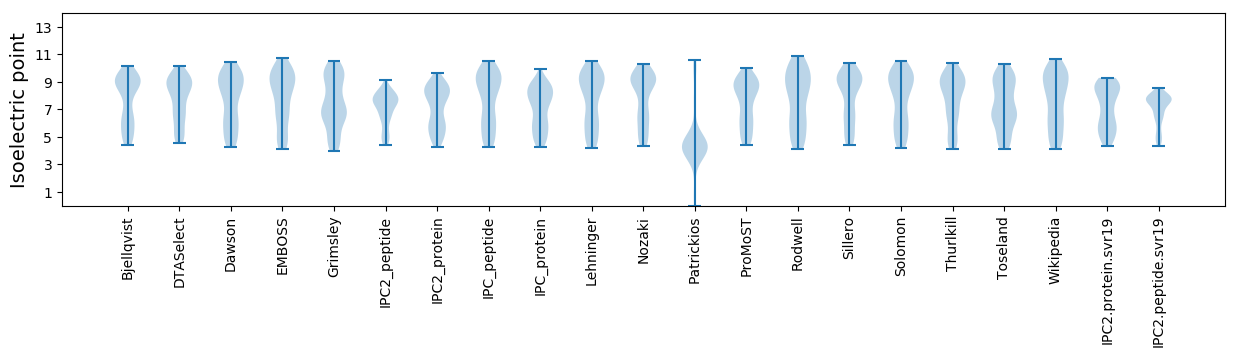
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q196W2|VF439_IIV3 Probable kinase 098L OS=Invertebrate iridescent virus 3 OX=345201 GN=IIV3-098L PE=3 SV=1
MM1 pKa = 7.53NDD3 pKa = 2.88VHH5 pKa = 7.45DD6 pKa = 4.38CLVWVNDD13 pKa = 4.43PYY15 pKa = 11.23FHH17 pKa = 7.6PISKK21 pKa = 10.41RR22 pKa = 11.84PIKK25 pKa = 10.7YY26 pKa = 9.82KK27 pKa = 10.33GPTWRR32 pKa = 11.84HH33 pKa = 4.3YY34 pKa = 10.83DD35 pKa = 3.65KK36 pKa = 11.33KK37 pKa = 10.89CDD39 pKa = 3.88LLGITGPKK47 pKa = 8.0ATKK50 pKa = 9.98SPSRR54 pKa = 11.84RR55 pKa = 11.84TTRR58 pKa = 11.84SPSPSRR64 pKa = 11.84RR65 pKa = 11.84TTRR68 pKa = 11.84SSPSRR73 pKa = 11.84RR74 pKa = 11.84TTRR77 pKa = 11.84SSPSRR82 pKa = 11.84RR83 pKa = 11.84TTRR86 pKa = 11.84SPSPSGRR93 pKa = 11.84RR94 pKa = 11.84KK95 pKa = 9.23QGGPAVYY102 pKa = 9.99CGNNALDD109 pKa = 4.11EE110 pKa = 4.48GLLDD114 pKa = 3.8GSKK117 pKa = 10.76VVGTRR122 pKa = 11.84YY123 pKa = 9.36QCLQKK128 pKa = 10.96GVAVGLNNPVLHH140 pKa = 6.81HH141 pKa = 6.65SPNYY145 pKa = 9.76QPIVDD150 pKa = 3.7AKK152 pKa = 9.33IYY154 pKa = 10.57CGTGSKK160 pKa = 10.61LPASKK165 pKa = 10.53LRR167 pKa = 11.84FGTPTEE173 pKa = 4.23CMGKK177 pKa = 10.28GYY179 pKa = 10.54QIGQNKK185 pKa = 9.3RR186 pKa = 11.84FQQSGLQQGPYY197 pKa = 9.18IWEE200 pKa = 3.6EE201 pKa = 4.0DD202 pKa = 3.07GWYY205 pKa = 10.41KK206 pKa = 10.62IIVPKK211 pKa = 10.75NN212 pKa = 3.12
MM1 pKa = 7.53NDD3 pKa = 2.88VHH5 pKa = 7.45DD6 pKa = 4.38CLVWVNDD13 pKa = 4.43PYY15 pKa = 11.23FHH17 pKa = 7.6PISKK21 pKa = 10.41RR22 pKa = 11.84PIKK25 pKa = 10.7YY26 pKa = 9.82KK27 pKa = 10.33GPTWRR32 pKa = 11.84HH33 pKa = 4.3YY34 pKa = 10.83DD35 pKa = 3.65KK36 pKa = 11.33KK37 pKa = 10.89CDD39 pKa = 3.88LLGITGPKK47 pKa = 8.0ATKK50 pKa = 9.98SPSRR54 pKa = 11.84RR55 pKa = 11.84TTRR58 pKa = 11.84SPSPSRR64 pKa = 11.84RR65 pKa = 11.84TTRR68 pKa = 11.84SSPSRR73 pKa = 11.84RR74 pKa = 11.84TTRR77 pKa = 11.84SSPSRR82 pKa = 11.84RR83 pKa = 11.84TTRR86 pKa = 11.84SPSPSGRR93 pKa = 11.84RR94 pKa = 11.84KK95 pKa = 9.23QGGPAVYY102 pKa = 9.99CGNNALDD109 pKa = 4.11EE110 pKa = 4.48GLLDD114 pKa = 3.8GSKK117 pKa = 10.76VVGTRR122 pKa = 11.84YY123 pKa = 9.36QCLQKK128 pKa = 10.96GVAVGLNNPVLHH140 pKa = 6.81HH141 pKa = 6.65SPNYY145 pKa = 9.76QPIVDD150 pKa = 3.7AKK152 pKa = 9.33IYY154 pKa = 10.57CGTGSKK160 pKa = 10.61LPASKK165 pKa = 10.53LRR167 pKa = 11.84FGTPTEE173 pKa = 4.23CMGKK177 pKa = 10.28GYY179 pKa = 10.54QIGQNKK185 pKa = 9.3RR186 pKa = 11.84FQQSGLQQGPYY197 pKa = 9.18IWEE200 pKa = 3.6EE201 pKa = 4.0DD202 pKa = 3.07GWYY205 pKa = 10.41KK206 pKa = 10.62IIVPKK211 pKa = 10.75NN212 pKa = 3.12
Molecular weight: 23.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
43365 |
60 |
1377 |
344.2 |
39.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.145 ± 0.192 | 1.783 ± 0.102 |
5.8 ± 0.119 | 5.654 ± 0.227 |
4.423 ± 0.152 | 4.983 ± 0.246 |
2.181 ± 0.131 | 5.938 ± 0.231 |
7.089 ± 0.238 | 8.982 ± 0.208 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.122 ± 0.086 | 5.313 ± 0.128 |
5.447 ± 0.245 | 4.381 ± 0.123 |
4.822 ± 0.181 | 7.059 ± 0.242 |
6.768 ± 0.205 | 6.874 ± 0.173 |
1.351 ± 0.088 | 3.886 ± 0.13 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
