
Shayang ascaridia galli virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betanemrhavirus; Shayang betanemrhavirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
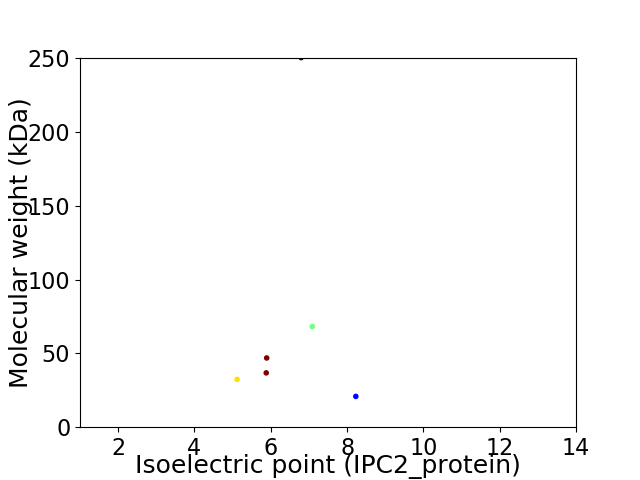
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KN28|A0A1L3KN28_9VIRU Putative nucleoprotein OS=Shayang ascaridia galli virus 2 OX=1923460 PE=4 SV=1
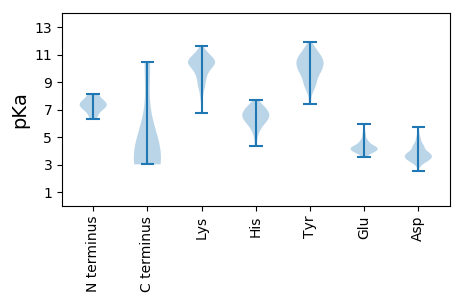
MM1 pKa = 8.14DD2 pKa = 5.69SNDD5 pKa = 3.5TLLDD9 pKa = 3.95GNEE12 pKa = 3.91LQEE15 pKa = 4.69AEE17 pKa = 4.13HH18 pKa = 6.65HH19 pKa = 6.54KK20 pKa = 11.17NSLLALAQDD29 pKa = 3.87TNFVSKK35 pKa = 9.64YY36 pKa = 8.33TLTSGDD42 pKa = 3.62FADD45 pKa = 4.99TEE47 pKa = 4.31DD48 pKa = 4.98PYY50 pKa = 11.88DD51 pKa = 3.65DD52 pKa = 4.37TRR54 pKa = 11.84FQLPQTPTSEE64 pKa = 4.35STTTAASYY72 pKa = 9.93PPRR75 pKa = 11.84TYY77 pKa = 9.98ATSSSDD83 pKa = 3.08HH84 pKa = 6.92SDD86 pKa = 3.67DD87 pKa = 5.54LRR89 pKa = 11.84HH90 pKa = 6.53KK91 pKa = 10.11YY92 pKa = 10.04INEE95 pKa = 3.61AAAALHH101 pKa = 6.27KK102 pKa = 10.76AGIPIDD108 pKa = 4.41SINQFLYY115 pKa = 9.81THH117 pKa = 7.47DD118 pKa = 3.77KK119 pKa = 10.67NKK121 pKa = 10.5AGFWEE126 pKa = 5.01GVVQCANLFLTKK138 pKa = 10.32SSSQALDD145 pKa = 4.75AIPLLHH151 pKa = 6.84SKK153 pKa = 9.93IDD155 pKa = 3.72SLQKK159 pKa = 10.82QMANLCEE166 pKa = 4.24LLTTNDD172 pKa = 3.3KK173 pKa = 10.06LTVQNHH179 pKa = 5.23NKK181 pKa = 10.17EE182 pKa = 4.59SVQSQQHH189 pKa = 5.91SGIQPEE195 pKa = 4.53PKK197 pKa = 8.74PQSVTPLEE205 pKa = 4.11IKK207 pKa = 10.81FNDD210 pKa = 3.62TTPLYY215 pKa = 10.87ARR217 pKa = 11.84EE218 pKa = 4.13AVTNLPIASACKK230 pKa = 9.93QYY232 pKa = 11.13LADD235 pKa = 3.66CMARR239 pKa = 11.84EE240 pKa = 4.41FIPWSFLHH248 pKa = 7.15AIHH251 pKa = 6.73NATSPNMKK259 pKa = 8.42RR260 pKa = 11.84TSYY263 pKa = 11.44LEE265 pKa = 3.82MIEE268 pKa = 3.96YY269 pKa = 8.53MRR271 pKa = 11.84SHH273 pKa = 6.46GMPPDD278 pKa = 3.44VYY280 pKa = 11.48DD281 pKa = 5.04FFIKK285 pKa = 10.08PWNN288 pKa = 3.54
MM1 pKa = 8.14DD2 pKa = 5.69SNDD5 pKa = 3.5TLLDD9 pKa = 3.95GNEE12 pKa = 3.91LQEE15 pKa = 4.69AEE17 pKa = 4.13HH18 pKa = 6.65HH19 pKa = 6.54KK20 pKa = 11.17NSLLALAQDD29 pKa = 3.87TNFVSKK35 pKa = 9.64YY36 pKa = 8.33TLTSGDD42 pKa = 3.62FADD45 pKa = 4.99TEE47 pKa = 4.31DD48 pKa = 4.98PYY50 pKa = 11.88DD51 pKa = 3.65DD52 pKa = 4.37TRR54 pKa = 11.84FQLPQTPTSEE64 pKa = 4.35STTTAASYY72 pKa = 9.93PPRR75 pKa = 11.84TYY77 pKa = 9.98ATSSSDD83 pKa = 3.08HH84 pKa = 6.92SDD86 pKa = 3.67DD87 pKa = 5.54LRR89 pKa = 11.84HH90 pKa = 6.53KK91 pKa = 10.11YY92 pKa = 10.04INEE95 pKa = 3.61AAAALHH101 pKa = 6.27KK102 pKa = 10.76AGIPIDD108 pKa = 4.41SINQFLYY115 pKa = 9.81THH117 pKa = 7.47DD118 pKa = 3.77KK119 pKa = 10.67NKK121 pKa = 10.5AGFWEE126 pKa = 5.01GVVQCANLFLTKK138 pKa = 10.32SSSQALDD145 pKa = 4.75AIPLLHH151 pKa = 6.84SKK153 pKa = 9.93IDD155 pKa = 3.72SLQKK159 pKa = 10.82QMANLCEE166 pKa = 4.24LLTTNDD172 pKa = 3.3KK173 pKa = 10.06LTVQNHH179 pKa = 5.23NKK181 pKa = 10.17EE182 pKa = 4.59SVQSQQHH189 pKa = 5.91SGIQPEE195 pKa = 4.53PKK197 pKa = 8.74PQSVTPLEE205 pKa = 4.11IKK207 pKa = 10.81FNDD210 pKa = 3.62TTPLYY215 pKa = 10.87ARR217 pKa = 11.84EE218 pKa = 4.13AVTNLPIASACKK230 pKa = 9.93QYY232 pKa = 11.13LADD235 pKa = 3.66CMARR239 pKa = 11.84EE240 pKa = 4.41FIPWSFLHH248 pKa = 7.15AIHH251 pKa = 6.73NATSPNMKK259 pKa = 8.42RR260 pKa = 11.84TSYY263 pKa = 11.44LEE265 pKa = 3.82MIEE268 pKa = 3.96YY269 pKa = 8.53MRR271 pKa = 11.84SHH273 pKa = 6.46GMPPDD278 pKa = 3.44VYY280 pKa = 11.48DD281 pKa = 5.04FFIKK285 pKa = 10.08PWNN288 pKa = 3.54
Molecular weight: 32.39 kDa
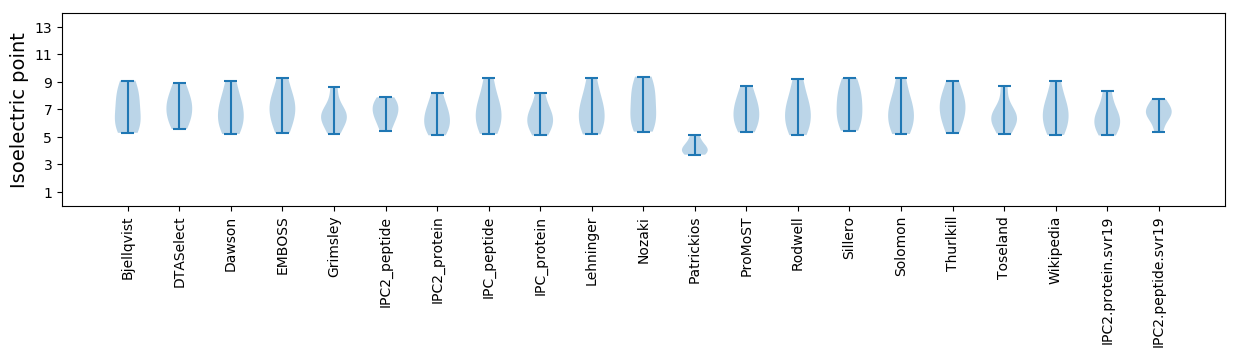
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KN19|A0A1L3KN19_9VIRU Uncharacterized protein OS=Shayang ascaridia galli virus 2 OX=1923460 PE=4 SV=1
MM1 pKa = 6.33TTYY4 pKa = 11.34LVMNMSGVIDD14 pKa = 3.85VSVAPPADD22 pKa = 3.47KK23 pKa = 11.19QEE25 pKa = 4.39TVDD28 pKa = 3.79IACQMVMAAITPPIPTEE45 pKa = 3.76LRR47 pKa = 11.84AAAARR52 pKa = 11.84ILCNKK57 pKa = 9.77LDD59 pKa = 3.76EE60 pKa = 4.69CRR62 pKa = 11.84PLQSTVITSNGKK74 pKa = 9.54VPGLRR79 pKa = 11.84YY80 pKa = 8.95PVLLDD85 pKa = 3.2GKK87 pKa = 10.32FAVKK91 pKa = 10.43LKK93 pKa = 10.43ADD95 pKa = 3.74RR96 pKa = 11.84PTRR99 pKa = 11.84DD100 pKa = 3.75FVSLHH105 pKa = 5.86CPQRR109 pKa = 11.84LDD111 pKa = 5.17NEE113 pKa = 4.52DD114 pKa = 4.37DD115 pKa = 3.88NNNSLFIHH123 pKa = 6.22LTLSAEE129 pKa = 4.02LGIINGVTANQLTITGRR146 pKa = 11.84HH147 pKa = 4.54IVPYY151 pKa = 10.05LGNDD155 pKa = 3.12EE156 pKa = 4.22RR157 pKa = 11.84FEE159 pKa = 5.23AIAAASIKK167 pKa = 10.33KK168 pKa = 10.46KK169 pKa = 10.57SDD171 pKa = 3.36TPKK174 pKa = 10.17SPIRR178 pKa = 11.84RR179 pKa = 11.84FAALFKK185 pKa = 10.74KK186 pKa = 10.39KK187 pKa = 9.85NHH189 pKa = 5.64NN190 pKa = 3.5
MM1 pKa = 6.33TTYY4 pKa = 11.34LVMNMSGVIDD14 pKa = 3.85VSVAPPADD22 pKa = 3.47KK23 pKa = 11.19QEE25 pKa = 4.39TVDD28 pKa = 3.79IACQMVMAAITPPIPTEE45 pKa = 3.76LRR47 pKa = 11.84AAAARR52 pKa = 11.84ILCNKK57 pKa = 9.77LDD59 pKa = 3.76EE60 pKa = 4.69CRR62 pKa = 11.84PLQSTVITSNGKK74 pKa = 9.54VPGLRR79 pKa = 11.84YY80 pKa = 8.95PVLLDD85 pKa = 3.2GKK87 pKa = 10.32FAVKK91 pKa = 10.43LKK93 pKa = 10.43ADD95 pKa = 3.74RR96 pKa = 11.84PTRR99 pKa = 11.84DD100 pKa = 3.75FVSLHH105 pKa = 5.86CPQRR109 pKa = 11.84LDD111 pKa = 5.17NEE113 pKa = 4.52DD114 pKa = 4.37DD115 pKa = 3.88NNNSLFIHH123 pKa = 6.22LTLSAEE129 pKa = 4.02LGIINGVTANQLTITGRR146 pKa = 11.84HH147 pKa = 4.54IVPYY151 pKa = 10.05LGNDD155 pKa = 3.12EE156 pKa = 4.22RR157 pKa = 11.84FEE159 pKa = 5.23AIAAASIKK167 pKa = 10.33KK168 pKa = 10.46KK169 pKa = 10.57SDD171 pKa = 3.36TPKK174 pKa = 10.17SPIRR178 pKa = 11.84RR179 pKa = 11.84FAALFKK185 pKa = 10.74KK186 pKa = 10.39KK187 pKa = 9.85NHH189 pKa = 5.64NN190 pKa = 3.5
Molecular weight: 20.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4061 |
190 |
2222 |
676.8 |
75.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.535 ± 0.636 | 2.241 ± 0.38 |
5.27 ± 0.417 | 5.319 ± 0.307 |
4.186 ± 0.376 | 4.432 ± 0.294 |
2.339 ± 0.31 | 5.639 ± 0.509 |
5.097 ± 0.281 | 10.81 ± 0.965 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.192 | 4.26 ± 0.582 |
5.023 ± 0.345 | 3.644 ± 0.468 |
5.294 ± 0.542 | 8.422 ± 0.39 |
6.722 ± 0.746 | 6.846 ± 0.578 |
1.551 ± 0.288 | 2.955 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
