
Myotis ricketti papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Treisiotapapillomavirus; Treisiotapapillomavirus 1
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
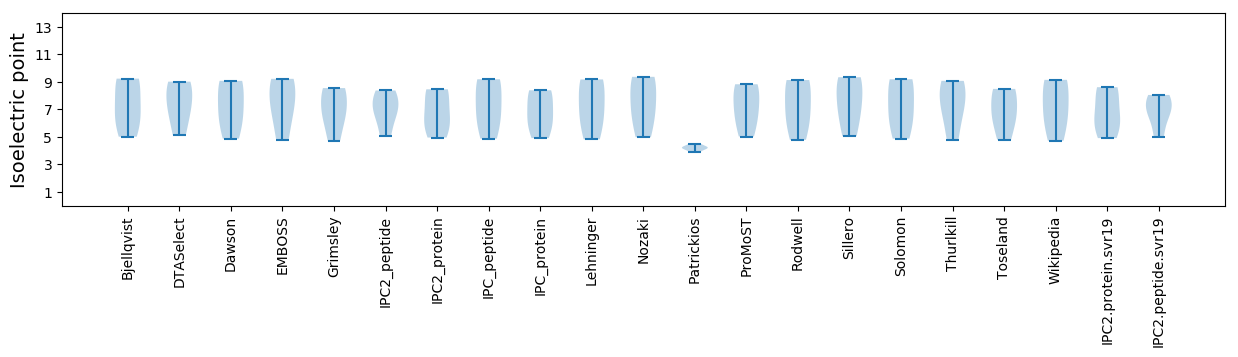
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|I3VR47|I3VR47_9PAPI Regulatory protein E2 OS=Myotis ricketti papillomavirus 1 OX=1195370 GN=E2 PE=3 SV=1
MM1 pKa = 7.75ANPPGNNGGTGCIYY15 pKa = 10.87LLDD18 pKa = 4.2EE19 pKa = 5.06AEE21 pKa = 4.74CDD23 pKa = 4.14DD24 pKa = 4.4GSSDD28 pKa = 3.49EE29 pKa = 4.94DD30 pKa = 3.75EE31 pKa = 4.72EE32 pKa = 4.45EE33 pKa = 4.46EE34 pKa = 5.06EE35 pKa = 4.38VGEE38 pKa = 4.93DD39 pKa = 3.86CVDD42 pKa = 4.81DD43 pKa = 4.12AACYY47 pKa = 9.56QGNTIALFVAQEE59 pKa = 3.84ASTDD63 pKa = 3.52EE64 pKa = 5.77AIIQSLKK71 pKa = 10.15RR72 pKa = 11.84KK73 pKa = 8.33YY74 pKa = 10.83VKK76 pKa = 9.83TPQQDD81 pKa = 3.47RR82 pKa = 11.84IRR84 pKa = 11.84EE85 pKa = 4.1IEE87 pKa = 4.06EE88 pKa = 4.21EE89 pKa = 3.65ISPRR93 pKa = 11.84LEE95 pKa = 4.6GIHH98 pKa = 7.0LEE100 pKa = 4.19THH102 pKa = 4.5SQKK105 pKa = 10.41VKK107 pKa = 10.72RR108 pKa = 11.84KK109 pKa = 9.71LFQANNAAKK118 pKa = 10.71GEE120 pKa = 4.16LDD122 pKa = 3.51VPLEE126 pKa = 4.3KK127 pKa = 10.44KK128 pKa = 10.31VATAGNLVEE137 pKa = 4.99VVAEE141 pKa = 4.33VHH143 pKa = 6.31CPPAGHH149 pKa = 7.46DD150 pKa = 3.5EE151 pKa = 4.62TVCSQKK157 pKa = 11.16SMEE160 pKa = 4.28GVEE163 pKa = 4.24EE164 pKa = 3.89QDD166 pKa = 4.13EE167 pKa = 5.09LGTCDD172 pKa = 3.15TGQEE176 pKa = 4.15AEE178 pKa = 4.29SALQNTEE185 pKa = 4.03PEE187 pKa = 4.28IEE189 pKa = 4.3CLSLEE194 pKa = 3.98ITEE197 pKa = 4.66PPVIPEE203 pKa = 4.19RR204 pKa = 11.84PWLDD208 pKa = 3.0GCTTEE213 pKa = 4.65QEE215 pKa = 3.88IAAAHH220 pKa = 6.15AKK222 pKa = 9.5AKK224 pKa = 10.38KK225 pKa = 9.45IAALKK230 pKa = 9.65LQEE233 pKa = 4.22KK234 pKa = 10.37AKK236 pKa = 11.04LPIQLLKK243 pKa = 9.86TGNRR247 pKa = 11.84KK248 pKa = 8.64AQILSHH254 pKa = 6.1CKK256 pKa = 8.48KK257 pKa = 8.91TFGISFIDD265 pKa = 3.11ITRR268 pKa = 11.84PFKK271 pKa = 11.0NDD273 pKa = 2.74NTVNEE278 pKa = 4.26TWVILSLYY286 pKa = 10.32LEE288 pKa = 4.58EE289 pKa = 4.52EE290 pKa = 4.34VADD293 pKa = 4.07ALLTVLEE300 pKa = 4.65PHH302 pKa = 6.91CKK304 pKa = 9.69CCFIISKK311 pKa = 10.27YY312 pKa = 10.53FGILKK317 pKa = 10.27SVLILIEE324 pKa = 4.03FQTAKK329 pKa = 10.32SRR331 pKa = 11.84KK332 pKa = 6.91TVLKK336 pKa = 10.81LMSSVMAVDD345 pKa = 4.17PEE347 pKa = 4.74AIIADD352 pKa = 3.97PPRR355 pKa = 11.84VRR357 pKa = 11.84SMPAALYY364 pKa = 7.98WYY366 pKa = 10.23KK367 pKa = 11.08VIAFEE372 pKa = 4.81KK373 pKa = 9.61AAQFGEE379 pKa = 4.13IPGWILTQVTLQHH392 pKa = 6.48SEE394 pKa = 3.96EE395 pKa = 4.52SEE397 pKa = 4.38LPFDD401 pKa = 4.98LSVMVQWAYY410 pKa = 11.31DD411 pKa = 3.45NDD413 pKa = 3.85YY414 pKa = 9.69DD415 pKa = 4.02TDD417 pKa = 3.88YY418 pKa = 11.47DD419 pKa = 4.03IAFEE423 pKa = 4.09YY424 pKa = 10.9AKK426 pKa = 10.75LATADD431 pKa = 3.57RR432 pKa = 11.84NARR435 pKa = 11.84AFLNSNCQAKK445 pKa = 9.54YY446 pKa = 11.01VKK448 pKa = 10.23DD449 pKa = 3.8CQTMVRR455 pKa = 11.84LYY457 pKa = 10.67RR458 pKa = 11.84KK459 pKa = 9.58AQMHH463 pKa = 5.98RR464 pKa = 11.84MSMGAWIKK472 pKa = 10.37EE473 pKa = 3.96RR474 pKa = 11.84SDD476 pKa = 4.72KK477 pKa = 10.78IDD479 pKa = 3.6GDD481 pKa = 4.02GDD483 pKa = 3.23WRR485 pKa = 11.84IIVHH489 pKa = 5.88FLRR492 pKa = 11.84YY493 pKa = 9.59QEE495 pKa = 4.29VEE497 pKa = 4.21FVSFMVKK504 pKa = 10.46FKK506 pKa = 11.11LFLKK510 pKa = 7.95GTPKK514 pKa = 10.42KK515 pKa = 10.59NCLLFYY521 pKa = 10.61GPPNTGKK528 pKa = 10.3SAFCMSLIKK537 pKa = 10.37FFAGRR542 pKa = 11.84VVSFMNARR550 pKa = 11.84SHH552 pKa = 7.32FWLQPLADD560 pKa = 4.26CRR562 pKa = 11.84VGLLDD567 pKa = 5.71DD568 pKa = 4.59ATDD571 pKa = 3.8ACWDD575 pKa = 3.74YY576 pKa = 11.46VDD578 pKa = 4.08TYY580 pKa = 10.47MRR582 pKa = 11.84SAFDD586 pKa = 3.57GNEE589 pKa = 3.25ISLDD593 pKa = 4.03CKK595 pKa = 10.61HH596 pKa = 6.61KK597 pKa = 11.14APIQLKK603 pKa = 10.0CPPLLVTSNTNILTSDD619 pKa = 2.78RR620 pKa = 11.84WKK622 pKa = 10.15YY623 pKa = 8.81LHH625 pKa = 7.03SRR627 pKa = 11.84VTVVKK632 pKa = 9.9FMNPFPLTQDD642 pKa = 3.07GQPRR646 pKa = 11.84YY647 pKa = 10.38NFQPKK652 pKa = 9.24DD653 pKa = 3.17WKK655 pKa = 11.13SFLSRR660 pKa = 11.84LWARR664 pKa = 11.84LDD666 pKa = 3.47LSEE669 pKa = 4.9PEE671 pKa = 4.16EE672 pKa = 4.51DD673 pKa = 5.4EE674 pKa = 4.62EE675 pKa = 5.38DD676 pKa = 3.13EE677 pKa = 5.0GYY679 pKa = 11.16NGDD682 pKa = 3.57SQEE685 pKa = 4.43KK686 pKa = 8.63FRR688 pKa = 11.84CTIRR692 pKa = 11.84EE693 pKa = 3.96NDD695 pKa = 3.13EE696 pKa = 4.07HH697 pKa = 7.89LL698 pKa = 4.27
MM1 pKa = 7.75ANPPGNNGGTGCIYY15 pKa = 10.87LLDD18 pKa = 4.2EE19 pKa = 5.06AEE21 pKa = 4.74CDD23 pKa = 4.14DD24 pKa = 4.4GSSDD28 pKa = 3.49EE29 pKa = 4.94DD30 pKa = 3.75EE31 pKa = 4.72EE32 pKa = 4.45EE33 pKa = 4.46EE34 pKa = 5.06EE35 pKa = 4.38VGEE38 pKa = 4.93DD39 pKa = 3.86CVDD42 pKa = 4.81DD43 pKa = 4.12AACYY47 pKa = 9.56QGNTIALFVAQEE59 pKa = 3.84ASTDD63 pKa = 3.52EE64 pKa = 5.77AIIQSLKK71 pKa = 10.15RR72 pKa = 11.84KK73 pKa = 8.33YY74 pKa = 10.83VKK76 pKa = 9.83TPQQDD81 pKa = 3.47RR82 pKa = 11.84IRR84 pKa = 11.84EE85 pKa = 4.1IEE87 pKa = 4.06EE88 pKa = 4.21EE89 pKa = 3.65ISPRR93 pKa = 11.84LEE95 pKa = 4.6GIHH98 pKa = 7.0LEE100 pKa = 4.19THH102 pKa = 4.5SQKK105 pKa = 10.41VKK107 pKa = 10.72RR108 pKa = 11.84KK109 pKa = 9.71LFQANNAAKK118 pKa = 10.71GEE120 pKa = 4.16LDD122 pKa = 3.51VPLEE126 pKa = 4.3KK127 pKa = 10.44KK128 pKa = 10.31VATAGNLVEE137 pKa = 4.99VVAEE141 pKa = 4.33VHH143 pKa = 6.31CPPAGHH149 pKa = 7.46DD150 pKa = 3.5EE151 pKa = 4.62TVCSQKK157 pKa = 11.16SMEE160 pKa = 4.28GVEE163 pKa = 4.24EE164 pKa = 3.89QDD166 pKa = 4.13EE167 pKa = 5.09LGTCDD172 pKa = 3.15TGQEE176 pKa = 4.15AEE178 pKa = 4.29SALQNTEE185 pKa = 4.03PEE187 pKa = 4.28IEE189 pKa = 4.3CLSLEE194 pKa = 3.98ITEE197 pKa = 4.66PPVIPEE203 pKa = 4.19RR204 pKa = 11.84PWLDD208 pKa = 3.0GCTTEE213 pKa = 4.65QEE215 pKa = 3.88IAAAHH220 pKa = 6.15AKK222 pKa = 9.5AKK224 pKa = 10.38KK225 pKa = 9.45IAALKK230 pKa = 9.65LQEE233 pKa = 4.22KK234 pKa = 10.37AKK236 pKa = 11.04LPIQLLKK243 pKa = 9.86TGNRR247 pKa = 11.84KK248 pKa = 8.64AQILSHH254 pKa = 6.1CKK256 pKa = 8.48KK257 pKa = 8.91TFGISFIDD265 pKa = 3.11ITRR268 pKa = 11.84PFKK271 pKa = 11.0NDD273 pKa = 2.74NTVNEE278 pKa = 4.26TWVILSLYY286 pKa = 10.32LEE288 pKa = 4.58EE289 pKa = 4.52EE290 pKa = 4.34VADD293 pKa = 4.07ALLTVLEE300 pKa = 4.65PHH302 pKa = 6.91CKK304 pKa = 9.69CCFIISKK311 pKa = 10.27YY312 pKa = 10.53FGILKK317 pKa = 10.27SVLILIEE324 pKa = 4.03FQTAKK329 pKa = 10.32SRR331 pKa = 11.84KK332 pKa = 6.91TVLKK336 pKa = 10.81LMSSVMAVDD345 pKa = 4.17PEE347 pKa = 4.74AIIADD352 pKa = 3.97PPRR355 pKa = 11.84VRR357 pKa = 11.84SMPAALYY364 pKa = 7.98WYY366 pKa = 10.23KK367 pKa = 11.08VIAFEE372 pKa = 4.81KK373 pKa = 9.61AAQFGEE379 pKa = 4.13IPGWILTQVTLQHH392 pKa = 6.48SEE394 pKa = 3.96EE395 pKa = 4.52SEE397 pKa = 4.38LPFDD401 pKa = 4.98LSVMVQWAYY410 pKa = 11.31DD411 pKa = 3.45NDD413 pKa = 3.85YY414 pKa = 9.69DD415 pKa = 4.02TDD417 pKa = 3.88YY418 pKa = 11.47DD419 pKa = 4.03IAFEE423 pKa = 4.09YY424 pKa = 10.9AKK426 pKa = 10.75LATADD431 pKa = 3.57RR432 pKa = 11.84NARR435 pKa = 11.84AFLNSNCQAKK445 pKa = 9.54YY446 pKa = 11.01VKK448 pKa = 10.23DD449 pKa = 3.8CQTMVRR455 pKa = 11.84LYY457 pKa = 10.67RR458 pKa = 11.84KK459 pKa = 9.58AQMHH463 pKa = 5.98RR464 pKa = 11.84MSMGAWIKK472 pKa = 10.37EE473 pKa = 3.96RR474 pKa = 11.84SDD476 pKa = 4.72KK477 pKa = 10.78IDD479 pKa = 3.6GDD481 pKa = 4.02GDD483 pKa = 3.23WRR485 pKa = 11.84IIVHH489 pKa = 5.88FLRR492 pKa = 11.84YY493 pKa = 9.59QEE495 pKa = 4.29VEE497 pKa = 4.21FVSFMVKK504 pKa = 10.46FKK506 pKa = 11.11LFLKK510 pKa = 7.95GTPKK514 pKa = 10.42KK515 pKa = 10.59NCLLFYY521 pKa = 10.61GPPNTGKK528 pKa = 10.3SAFCMSLIKK537 pKa = 10.37FFAGRR542 pKa = 11.84VVSFMNARR550 pKa = 11.84SHH552 pKa = 7.32FWLQPLADD560 pKa = 4.26CRR562 pKa = 11.84VGLLDD567 pKa = 5.71DD568 pKa = 4.59ATDD571 pKa = 3.8ACWDD575 pKa = 3.74YY576 pKa = 11.46VDD578 pKa = 4.08TYY580 pKa = 10.47MRR582 pKa = 11.84SAFDD586 pKa = 3.57GNEE589 pKa = 3.25ISLDD593 pKa = 4.03CKK595 pKa = 10.61HH596 pKa = 6.61KK597 pKa = 11.14APIQLKK603 pKa = 10.0CPPLLVTSNTNILTSDD619 pKa = 2.78RR620 pKa = 11.84WKK622 pKa = 10.15YY623 pKa = 8.81LHH625 pKa = 7.03SRR627 pKa = 11.84VTVVKK632 pKa = 9.9FMNPFPLTQDD642 pKa = 3.07GQPRR646 pKa = 11.84YY647 pKa = 10.38NFQPKK652 pKa = 9.24DD653 pKa = 3.17WKK655 pKa = 11.13SFLSRR660 pKa = 11.84LWARR664 pKa = 11.84LDD666 pKa = 3.47LSEE669 pKa = 4.9PEE671 pKa = 4.16EE672 pKa = 4.51DD673 pKa = 5.4EE674 pKa = 4.62EE675 pKa = 5.38DD676 pKa = 3.13EE677 pKa = 5.0GYY679 pKa = 11.16NGDD682 pKa = 3.57SQEE685 pKa = 4.43KK686 pKa = 8.63FRR688 pKa = 11.84CTIRR692 pKa = 11.84EE693 pKa = 3.96NDD695 pKa = 3.13EE696 pKa = 4.07HH697 pKa = 7.89LL698 pKa = 4.27
Molecular weight: 79.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3VR46|I3VR46_9PAPI Replication protein E1 OS=Myotis ricketti papillomavirus 1 OX=1195370 GN=E1 PE=3 SV=1
MM1 pKa = 7.27MIVVNYY7 pKa = 9.4YY8 pKa = 10.34DD9 pKa = 3.44KK10 pKa = 10.97KK11 pKa = 10.68GKK13 pKa = 9.28FRR15 pKa = 11.84RR16 pKa = 11.84DD17 pKa = 3.13RR18 pKa = 11.84SRR20 pKa = 11.84SQYY23 pKa = 10.64ISIRR27 pKa = 11.84PIRR30 pKa = 11.84GASMGSVTKK39 pKa = 10.76RR40 pKa = 11.84EE41 pKa = 4.15DD42 pKa = 3.13GEE44 pKa = 4.04YY45 pKa = 10.16RR46 pKa = 11.84PRR48 pKa = 11.84SLRR51 pKa = 11.84EE52 pKa = 3.62LMVDD56 pKa = 3.95MGLQYY61 pKa = 9.49QTILLSCVYY70 pKa = 10.04CSKK73 pKa = 9.72TLEE76 pKa = 4.03VSEE79 pKa = 5.0LVTFSVKK86 pKa = 10.31DD87 pKa = 3.39LQIIWRR93 pKa = 11.84GGFPYY98 pKa = 10.25GVCAFCLEE106 pKa = 4.46FNLQIQYY113 pKa = 10.19LRR115 pKa = 11.84HH116 pKa = 5.75HH117 pKa = 6.81RR118 pKa = 11.84RR119 pKa = 11.84SASIKK124 pKa = 8.35TVEE127 pKa = 4.58EE128 pKa = 3.83EE129 pKa = 4.0LQRR132 pKa = 11.84PVGNLNLRR140 pKa = 11.84CFVCAIKK147 pKa = 10.55LHH149 pKa = 5.45STDD152 pKa = 3.41YY153 pKa = 10.58ATMEE157 pKa = 4.05EE158 pKa = 3.93QGLNLKK164 pKa = 10.09YY165 pKa = 10.25IAGQWRR171 pKa = 11.84GKK173 pKa = 10.83CMICIGAVEE182 pKa = 4.43GVV184 pKa = 3.21
MM1 pKa = 7.27MIVVNYY7 pKa = 9.4YY8 pKa = 10.34DD9 pKa = 3.44KK10 pKa = 10.97KK11 pKa = 10.68GKK13 pKa = 9.28FRR15 pKa = 11.84RR16 pKa = 11.84DD17 pKa = 3.13RR18 pKa = 11.84SRR20 pKa = 11.84SQYY23 pKa = 10.64ISIRR27 pKa = 11.84PIRR30 pKa = 11.84GASMGSVTKK39 pKa = 10.76RR40 pKa = 11.84EE41 pKa = 4.15DD42 pKa = 3.13GEE44 pKa = 4.04YY45 pKa = 10.16RR46 pKa = 11.84PRR48 pKa = 11.84SLRR51 pKa = 11.84EE52 pKa = 3.62LMVDD56 pKa = 3.95MGLQYY61 pKa = 9.49QTILLSCVYY70 pKa = 10.04CSKK73 pKa = 9.72TLEE76 pKa = 4.03VSEE79 pKa = 5.0LVTFSVKK86 pKa = 10.31DD87 pKa = 3.39LQIIWRR93 pKa = 11.84GGFPYY98 pKa = 10.25GVCAFCLEE106 pKa = 4.46FNLQIQYY113 pKa = 10.19LRR115 pKa = 11.84HH116 pKa = 5.75HH117 pKa = 6.81RR118 pKa = 11.84RR119 pKa = 11.84SASIKK124 pKa = 8.35TVEE127 pKa = 4.58EE128 pKa = 3.83EE129 pKa = 4.0LQRR132 pKa = 11.84PVGNLNLRR140 pKa = 11.84CFVCAIKK147 pKa = 10.55LHH149 pKa = 5.45STDD152 pKa = 3.41YY153 pKa = 10.58ATMEE157 pKa = 4.05EE158 pKa = 3.93QGLNLKK164 pKa = 10.09YY165 pKa = 10.25IAGQWRR171 pKa = 11.84GKK173 pKa = 10.83CMICIGAVEE182 pKa = 4.43GVV184 pKa = 3.21
Molecular weight: 21.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2174 |
184 |
698 |
434.8 |
48.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.808 ± 0.558 | 2.162 ± 0.48 |
5.888 ± 0.448 | 6.348 ± 0.917 |
4.232 ± 0.492 | 6.118 ± 0.643 |
2.024 ± 0.099 | 5.152 ± 0.482 |
5.566 ± 0.629 | 8.05 ± 0.331 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.07 ± 0.32 | 3.588 ± 0.416 |
6.348 ± 0.861 | 4.324 ± 0.316 |
5.888 ± 0.582 | 7.498 ± 0.738 |
6.946 ± 0.787 | 5.934 ± 0.261 |
1.334 ± 0.321 | 3.726 ± 0.33 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
