
Pseudomonas oryzae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
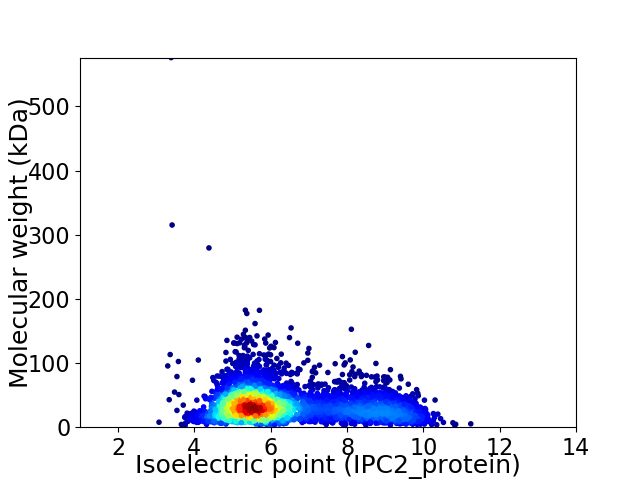
Virtual 2D-PAGE plot for 4103 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1S471|A0A1H1S471_9PSED 3-oxoacyl-[acyl-carrier-protein] synthase I OS=Pseudomonas oryzae OX=1392877 GN=SAMN05216221_1799 PE=3 SV=1
MM1 pKa = 7.1HH2 pKa = 6.51THH4 pKa = 6.67KK5 pKa = 10.55FLRR8 pKa = 11.84TLPRR12 pKa = 11.84SLLSAAVVAGIVTAAGVFAVDD33 pKa = 4.25GNPPGLFEE41 pKa = 6.54LDD43 pKa = 3.62GNTVDD48 pKa = 4.59VGGTPGDD55 pKa = 3.47DD56 pKa = 2.94WGSIFTGTNLGTPAASTGILADD78 pKa = 3.92PAPLTIYY85 pKa = 8.81TQGGSKK91 pKa = 10.42DD92 pKa = 3.56INDD95 pKa = 3.38VSEE98 pKa = 3.88WRR100 pKa = 11.84HH101 pKa = 4.73TNGNVPDD108 pKa = 4.94KK109 pKa = 11.4DD110 pKa = 4.92DD111 pKa = 3.37ITNAYY116 pKa = 8.52AAGYY120 pKa = 9.33INPNNVSHH128 pKa = 6.84NGQVVHH134 pKa = 5.96QAGDD138 pKa = 3.86LIIYY142 pKa = 9.61FGLDD146 pKa = 2.75RR147 pKa = 11.84YY148 pKa = 10.89ANNGDD153 pKa = 3.39AFAGFWFFQDD163 pKa = 4.93DD164 pKa = 3.84VGLGPNGTFQGVHH177 pKa = 5.1TARR180 pKa = 11.84DD181 pKa = 3.33GDD183 pKa = 3.61QRR185 pKa = 11.84GDD187 pKa = 3.57LLVLVEE193 pKa = 4.33YY194 pKa = 9.55PQGANAQPEE203 pKa = 4.43IKK205 pKa = 10.47VYY207 pKa = 10.24EE208 pKa = 4.34WDD210 pKa = 4.29PADD213 pKa = 4.25ADD215 pKa = 3.89NDD217 pKa = 3.8NVADD221 pKa = 4.26NLDD224 pKa = 4.28EE225 pKa = 5.12IYY227 pKa = 11.03SSTNAKK233 pKa = 10.11CDD235 pKa = 3.33GAGNKK240 pKa = 8.27LACAITNVNNLTNEE254 pKa = 4.27PAWDD258 pKa = 3.89YY259 pKa = 10.52TPKK262 pKa = 10.54SGPASDD268 pKa = 4.41LPKK271 pKa = 10.64EE272 pKa = 4.32SFFEE276 pKa = 4.09GGINVTRR283 pKa = 11.84LLGGATPCFSSFLAEE298 pKa = 4.17TRR300 pKa = 11.84SSRR303 pKa = 11.84SEE305 pKa = 3.7TAQLKK310 pKa = 10.88DD311 pKa = 3.67FVLGNFDD318 pKa = 3.78LCSIAVTKK326 pKa = 9.93TCEE329 pKa = 4.6AEE331 pKa = 4.83VNDD334 pKa = 3.72QDD336 pKa = 3.84GGNSVVVNFSGEE348 pKa = 4.04VTNDD352 pKa = 2.85GGLALNNVTVTDD364 pKa = 4.19DD365 pKa = 3.3MGTPGDD371 pKa = 3.82TGDD374 pKa = 3.86DD375 pKa = 3.19QVVFGPATLQPGEE388 pKa = 4.15TQPYY392 pKa = 7.61SGSYY396 pKa = 7.34EE397 pKa = 4.04TTAIPATDD405 pKa = 3.29QVTATGSRR413 pKa = 11.84GTATVEE419 pKa = 3.74ATADD423 pKa = 3.77ATCSPTVSPAITVDD437 pKa = 3.84KK438 pKa = 10.7FCSATLKK445 pKa = 10.65EE446 pKa = 4.85DD447 pKa = 3.36GSGVDD452 pKa = 3.32VLFNGTVTNTGNVALEE468 pKa = 4.17NVTVVDD474 pKa = 4.83DD475 pKa = 5.1NGTADD480 pKa = 4.16PGDD483 pKa = 4.23DD484 pKa = 3.66VTVLGPVTLNAGASMPYY501 pKa = 9.96NGGFGVTGSNTSTDD515 pKa = 3.33HH516 pKa = 6.03VVASGTDD523 pKa = 3.16VLTDD527 pKa = 3.46VEE529 pKa = 4.53VSDD532 pKa = 4.03TAEE535 pKa = 4.17ATCLADD541 pKa = 3.71TNPSILLAKK550 pKa = 10.12QCTATVNSSGDD561 pKa = 3.71GIDD564 pKa = 3.41VSFTGSVTNNGNVILNNVTVIDD586 pKa = 4.68DD587 pKa = 3.78NGTPGNTGDD596 pKa = 5.53DD597 pKa = 3.38VTLIDD602 pKa = 4.01NATLAVGASLPFSGNFSGSGSSSTDD627 pKa = 3.16VVSATASDD635 pKa = 4.04ALTGDD640 pKa = 3.8VVSDD644 pKa = 3.89TEE646 pKa = 4.39TATCAADD653 pKa = 3.57VNPAILVSKK662 pKa = 10.59QCTATVNSAGTAIDD676 pKa = 3.41VLFIGSVTNTGNVVLQNVNVVDD698 pKa = 5.08DD699 pKa = 4.64NGTPGNSGDD708 pKa = 4.32DD709 pKa = 3.38VTLISNATLGVGASLPFNGNFSSSSASSTDD739 pKa = 3.12VVTATGVDD747 pKa = 3.7MLTSQAVSDD756 pKa = 4.37TEE758 pKa = 4.41TASCAADD765 pKa = 4.24LNPSIAVTKK774 pKa = 10.5QCTDD778 pKa = 2.94AMAHH782 pKa = 6.0DD783 pKa = 4.19QPILFNGTVSNTGNVALLGVTVVDD807 pKa = 5.25DD808 pKa = 4.43NGTPADD814 pKa = 4.38PLDD817 pKa = 4.21DD818 pKa = 3.57QTFNLGDD825 pKa = 4.06LAPGASANYY834 pKa = 9.1NGSYY838 pKa = 10.69SPGTGTWTNTVEE850 pKa = 4.32ANANGALEE858 pKa = 4.48TGSFTATASATCDD871 pKa = 3.52VPPPPPEE878 pKa = 4.48FEE880 pKa = 4.29GCTPGFWKK888 pKa = 10.5NSPGSWVGYY897 pKa = 9.83SPNQLVGSVFSLPSGVLNNQLGDD920 pKa = 3.88DD921 pKa = 4.11TLMEE925 pKa = 4.51ALGYY929 pKa = 9.81PGGTNLVGAAQILLRR944 pKa = 11.84AAVASLLNAGHH955 pKa = 7.48PDD957 pKa = 2.91VDD959 pKa = 4.67FPLTTSEE966 pKa = 5.74VITQVNAALATKK978 pKa = 10.55DD979 pKa = 3.17RR980 pKa = 11.84GTILALASTLDD991 pKa = 3.58GYY993 pKa = 12.05NNLGCDD999 pKa = 4.05LPNDD1003 pKa = 3.53NSFF1006 pKa = 3.53
MM1 pKa = 7.1HH2 pKa = 6.51THH4 pKa = 6.67KK5 pKa = 10.55FLRR8 pKa = 11.84TLPRR12 pKa = 11.84SLLSAAVVAGIVTAAGVFAVDD33 pKa = 4.25GNPPGLFEE41 pKa = 6.54LDD43 pKa = 3.62GNTVDD48 pKa = 4.59VGGTPGDD55 pKa = 3.47DD56 pKa = 2.94WGSIFTGTNLGTPAASTGILADD78 pKa = 3.92PAPLTIYY85 pKa = 8.81TQGGSKK91 pKa = 10.42DD92 pKa = 3.56INDD95 pKa = 3.38VSEE98 pKa = 3.88WRR100 pKa = 11.84HH101 pKa = 4.73TNGNVPDD108 pKa = 4.94KK109 pKa = 11.4DD110 pKa = 4.92DD111 pKa = 3.37ITNAYY116 pKa = 8.52AAGYY120 pKa = 9.33INPNNVSHH128 pKa = 6.84NGQVVHH134 pKa = 5.96QAGDD138 pKa = 3.86LIIYY142 pKa = 9.61FGLDD146 pKa = 2.75RR147 pKa = 11.84YY148 pKa = 10.89ANNGDD153 pKa = 3.39AFAGFWFFQDD163 pKa = 4.93DD164 pKa = 3.84VGLGPNGTFQGVHH177 pKa = 5.1TARR180 pKa = 11.84DD181 pKa = 3.33GDD183 pKa = 3.61QRR185 pKa = 11.84GDD187 pKa = 3.57LLVLVEE193 pKa = 4.33YY194 pKa = 9.55PQGANAQPEE203 pKa = 4.43IKK205 pKa = 10.47VYY207 pKa = 10.24EE208 pKa = 4.34WDD210 pKa = 4.29PADD213 pKa = 4.25ADD215 pKa = 3.89NDD217 pKa = 3.8NVADD221 pKa = 4.26NLDD224 pKa = 4.28EE225 pKa = 5.12IYY227 pKa = 11.03SSTNAKK233 pKa = 10.11CDD235 pKa = 3.33GAGNKK240 pKa = 8.27LACAITNVNNLTNEE254 pKa = 4.27PAWDD258 pKa = 3.89YY259 pKa = 10.52TPKK262 pKa = 10.54SGPASDD268 pKa = 4.41LPKK271 pKa = 10.64EE272 pKa = 4.32SFFEE276 pKa = 4.09GGINVTRR283 pKa = 11.84LLGGATPCFSSFLAEE298 pKa = 4.17TRR300 pKa = 11.84SSRR303 pKa = 11.84SEE305 pKa = 3.7TAQLKK310 pKa = 10.88DD311 pKa = 3.67FVLGNFDD318 pKa = 3.78LCSIAVTKK326 pKa = 9.93TCEE329 pKa = 4.6AEE331 pKa = 4.83VNDD334 pKa = 3.72QDD336 pKa = 3.84GGNSVVVNFSGEE348 pKa = 4.04VTNDD352 pKa = 2.85GGLALNNVTVTDD364 pKa = 4.19DD365 pKa = 3.3MGTPGDD371 pKa = 3.82TGDD374 pKa = 3.86DD375 pKa = 3.19QVVFGPATLQPGEE388 pKa = 4.15TQPYY392 pKa = 7.61SGSYY396 pKa = 7.34EE397 pKa = 4.04TTAIPATDD405 pKa = 3.29QVTATGSRR413 pKa = 11.84GTATVEE419 pKa = 3.74ATADD423 pKa = 3.77ATCSPTVSPAITVDD437 pKa = 3.84KK438 pKa = 10.7FCSATLKK445 pKa = 10.65EE446 pKa = 4.85DD447 pKa = 3.36GSGVDD452 pKa = 3.32VLFNGTVTNTGNVALEE468 pKa = 4.17NVTVVDD474 pKa = 4.83DD475 pKa = 5.1NGTADD480 pKa = 4.16PGDD483 pKa = 4.23DD484 pKa = 3.66VTVLGPVTLNAGASMPYY501 pKa = 9.96NGGFGVTGSNTSTDD515 pKa = 3.33HH516 pKa = 6.03VVASGTDD523 pKa = 3.16VLTDD527 pKa = 3.46VEE529 pKa = 4.53VSDD532 pKa = 4.03TAEE535 pKa = 4.17ATCLADD541 pKa = 3.71TNPSILLAKK550 pKa = 10.12QCTATVNSSGDD561 pKa = 3.71GIDD564 pKa = 3.41VSFTGSVTNNGNVILNNVTVIDD586 pKa = 4.68DD587 pKa = 3.78NGTPGNTGDD596 pKa = 5.53DD597 pKa = 3.38VTLIDD602 pKa = 4.01NATLAVGASLPFSGNFSGSGSSSTDD627 pKa = 3.16VVSATASDD635 pKa = 4.04ALTGDD640 pKa = 3.8VVSDD644 pKa = 3.89TEE646 pKa = 4.39TATCAADD653 pKa = 3.57VNPAILVSKK662 pKa = 10.59QCTATVNSAGTAIDD676 pKa = 3.41VLFIGSVTNTGNVVLQNVNVVDD698 pKa = 5.08DD699 pKa = 4.64NGTPGNSGDD708 pKa = 4.32DD709 pKa = 3.38VTLISNATLGVGASLPFNGNFSSSSASSTDD739 pKa = 3.12VVTATGVDD747 pKa = 3.7MLTSQAVSDD756 pKa = 4.37TEE758 pKa = 4.41TASCAADD765 pKa = 4.24LNPSIAVTKK774 pKa = 10.5QCTDD778 pKa = 2.94AMAHH782 pKa = 6.0DD783 pKa = 4.19QPILFNGTVSNTGNVALLGVTVVDD807 pKa = 5.25DD808 pKa = 4.43NGTPADD814 pKa = 4.38PLDD817 pKa = 4.21DD818 pKa = 3.57QTFNLGDD825 pKa = 4.06LAPGASANYY834 pKa = 9.1NGSYY838 pKa = 10.69SPGTGTWTNTVEE850 pKa = 4.32ANANGALEE858 pKa = 4.48TGSFTATASATCDD871 pKa = 3.52VPPPPPEE878 pKa = 4.48FEE880 pKa = 4.29GCTPGFWKK888 pKa = 10.5NSPGSWVGYY897 pKa = 9.83SPNQLVGSVFSLPSGVLNNQLGDD920 pKa = 3.88DD921 pKa = 4.11TLMEE925 pKa = 4.51ALGYY929 pKa = 9.81PGGTNLVGAAQILLRR944 pKa = 11.84AAVASLLNAGHH955 pKa = 7.48PDD957 pKa = 2.91VDD959 pKa = 4.67FPLTTSEE966 pKa = 5.74VITQVNAALATKK978 pKa = 10.55DD979 pKa = 3.17RR980 pKa = 11.84GTILALASTLDD991 pKa = 3.58GYY993 pKa = 12.05NNLGCDD999 pKa = 4.05LPNDD1003 pKa = 3.53NSFF1006 pKa = 3.53
Molecular weight: 102.44 kDa
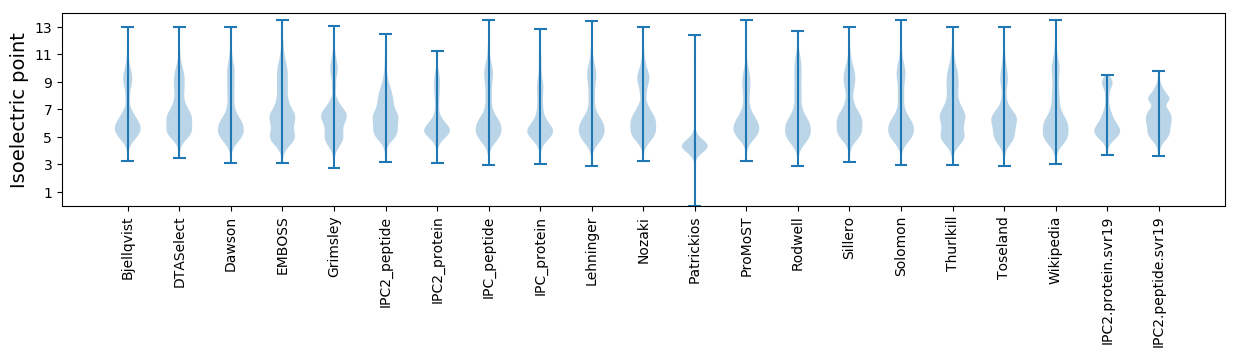
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1XPY9|A0A1H1XPY9_9PSED Beta sliding clamp OS=Pseudomonas oryzae OX=1392877 GN=SAMN05216221_3494 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 9.11RR41 pKa = 11.84LTTVV45 pKa = 2.62
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 9.11RR41 pKa = 11.84LTTVV45 pKa = 2.62
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1362556 |
29 |
5777 |
332.1 |
36.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.221 ± 0.054 | 1.09 ± 0.014 |
5.259 ± 0.034 | 6.101 ± 0.04 |
3.457 ± 0.023 | 8.345 ± 0.047 |
2.31 ± 0.022 | 4.326 ± 0.029 |
2.879 ± 0.035 | 12.352 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.138 ± 0.022 | 2.56 ± 0.027 |
4.988 ± 0.028 | 4.359 ± 0.033 |
7.193 ± 0.048 | 5.273 ± 0.034 |
4.312 ± 0.05 | 7.005 ± 0.034 |
1.424 ± 0.016 | 2.407 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
