
Hubei diptera virus 13
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.9
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
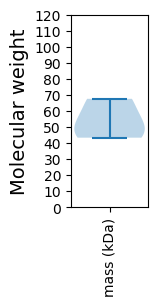
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEL1|A0A1L3KEL1_9VIRU RNA-directed RNA polymerase OS=Hubei diptera virus 13 OX=1922874 PE=4 SV=1
MM1 pKa = 7.45SVEE4 pKa = 4.21DD5 pKa = 4.08LVDD8 pKa = 4.34FVGEE12 pKa = 4.06TARR15 pKa = 11.84LEE17 pKa = 4.31VEE19 pKa = 4.39SQVGDD24 pKa = 3.56TLFSRR29 pKa = 11.84NGLKK33 pKa = 10.12EE34 pKa = 3.8RR35 pKa = 11.84AKK37 pKa = 10.62KK38 pKa = 9.82IALSATLLCARR49 pKa = 11.84ALLGTVLVLCCGAWATAAVMALVSVAKK76 pKa = 10.15HH77 pKa = 5.29AMWYY81 pKa = 10.21QYY83 pKa = 11.59DD84 pKa = 3.86SLYY87 pKa = 11.18KK88 pKa = 10.65LIVQSFVPEE97 pKa = 4.39PEE99 pKa = 4.28PEE101 pKa = 3.88PDD103 pKa = 3.61YY104 pKa = 11.17FSGLISDD111 pKa = 3.84VTALVKK117 pKa = 10.56RR118 pKa = 11.84VDD120 pKa = 3.73LTLLSLLLVVLMILASIWISMGYY143 pKa = 9.65VYY145 pKa = 11.22YY146 pKa = 11.01SMRR149 pKa = 11.84RR150 pKa = 11.84VSYY153 pKa = 9.82KK154 pKa = 10.19VRR156 pKa = 11.84GIQYY160 pKa = 9.59EE161 pKa = 3.97AMQAGSEE168 pKa = 4.11FRR170 pKa = 11.84EE171 pKa = 4.49GEE173 pKa = 4.08VPKK176 pKa = 10.65FQVRR180 pKa = 11.84VLLPGVFVNSHH191 pKa = 4.53QGYY194 pKa = 9.5GIRR197 pKa = 11.84VEE199 pKa = 3.97NWLVLPTHH207 pKa = 6.38VLTGSEE213 pKa = 3.84ILLEE217 pKa = 4.28GSRR220 pKa = 11.84GRR222 pKa = 11.84VVIPTDD228 pKa = 3.52HH229 pKa = 6.92EE230 pKa = 4.27PSKK233 pKa = 10.43LHH235 pKa = 7.04PDD237 pKa = 3.25VAYY240 pKa = 10.76VWLEE244 pKa = 3.67DD245 pKa = 4.13SQWTTLGTRR254 pKa = 11.84SAKK257 pKa = 9.62TIPITSCHH265 pKa = 5.28VRR267 pKa = 11.84CTGQKK272 pKa = 8.49GYY274 pKa = 11.99SMGTVNKK281 pKa = 9.18TNIIGVLRR289 pKa = 11.84YY290 pKa = 9.93SGSTVPGMSGAAYY303 pKa = 8.9VAQDD307 pKa = 3.55RR308 pKa = 11.84VVGMHH313 pKa = 6.78TGASMEE319 pKa = 4.28KK320 pKa = 9.88NLGVAAALFVNEE332 pKa = 4.62LKK334 pKa = 10.9KK335 pKa = 10.69KK336 pKa = 9.83IKK338 pKa = 10.6NEE340 pKa = 4.28AIFLPSSNNSEE351 pKa = 3.93DD352 pKa = 3.51AAYY355 pKa = 10.69QMKK358 pKa = 10.44SFKK361 pKa = 8.53NTKK364 pKa = 8.7WDD366 pKa = 3.57AKK368 pKa = 10.57LLEE371 pKa = 4.55ALAEE375 pKa = 4.32NADD378 pKa = 4.18DD379 pKa = 4.28MDD381 pKa = 4.65LGSWGGNVDD390 pKa = 6.25FDD392 pKa = 5.12PDD394 pKa = 3.4WKK396 pKa = 11.08AEE398 pKa = 4.13SAEE401 pKa = 4.19EE402 pKa = 3.93PKK404 pKa = 10.51KK405 pKa = 10.76QNPIEE410 pKa = 4.14LATLIMKK417 pKa = 8.52QHH419 pKa = 6.05GPSGEE424 pKa = 4.02EE425 pKa = 3.7TGFSLLNTKK434 pKa = 9.01VLEE437 pKa = 4.94DD438 pKa = 3.39IKK440 pKa = 11.06DD441 pKa = 3.59LQEE444 pKa = 3.88RR445 pKa = 11.84LTIVEE450 pKa = 4.69EE451 pKa = 4.29YY452 pKa = 10.38IKK454 pKa = 10.21TKK456 pKa = 10.11ISKK459 pKa = 10.1KK460 pKa = 10.14KK461 pKa = 8.11EE462 pKa = 3.68PKK464 pKa = 10.18PIVTYY469 pKa = 10.57DD470 pKa = 3.63CEE472 pKa = 4.11EE473 pKa = 4.69CGAKK477 pKa = 7.53TTSEE481 pKa = 4.05SDD483 pKa = 3.51LTKK486 pKa = 10.67HH487 pKa = 4.98QAKK490 pKa = 9.66HH491 pKa = 5.52VKK493 pKa = 9.26HH494 pKa = 6.24KK495 pKa = 11.27CEE497 pKa = 3.96ICDD500 pKa = 3.45ITCQTALRR508 pKa = 11.84LANHH512 pKa = 6.29VANAHH517 pKa = 5.43RR518 pKa = 11.84TAKK521 pKa = 10.39PEE523 pKa = 3.76SAYY526 pKa = 10.17PGDD529 pKa = 3.52ARR531 pKa = 11.84VVVQTAPEE539 pKa = 4.12TQAFLDD545 pKa = 4.0PRR547 pKa = 11.84SNSPKK552 pKa = 9.9RR553 pKa = 11.84KK554 pKa = 8.52NRR556 pKa = 11.84SSKK559 pKa = 10.23RR560 pKa = 11.84SSSTSSKK567 pKa = 10.59KK568 pKa = 9.86QDD570 pKa = 3.33SLPWEE575 pKa = 4.07KK576 pKa = 10.72LLYY579 pKa = 10.78QMVQSQNRR587 pKa = 11.84LEE589 pKa = 4.15EE590 pKa = 4.07YY591 pKa = 10.42LRR593 pKa = 11.84SEE595 pKa = 4.53RR596 pKa = 11.84RR597 pKa = 11.84AMAGPASATQPKK609 pKa = 9.85
MM1 pKa = 7.45SVEE4 pKa = 4.21DD5 pKa = 4.08LVDD8 pKa = 4.34FVGEE12 pKa = 4.06TARR15 pKa = 11.84LEE17 pKa = 4.31VEE19 pKa = 4.39SQVGDD24 pKa = 3.56TLFSRR29 pKa = 11.84NGLKK33 pKa = 10.12EE34 pKa = 3.8RR35 pKa = 11.84AKK37 pKa = 10.62KK38 pKa = 9.82IALSATLLCARR49 pKa = 11.84ALLGTVLVLCCGAWATAAVMALVSVAKK76 pKa = 10.15HH77 pKa = 5.29AMWYY81 pKa = 10.21QYY83 pKa = 11.59DD84 pKa = 3.86SLYY87 pKa = 11.18KK88 pKa = 10.65LIVQSFVPEE97 pKa = 4.39PEE99 pKa = 4.28PEE101 pKa = 3.88PDD103 pKa = 3.61YY104 pKa = 11.17FSGLISDD111 pKa = 3.84VTALVKK117 pKa = 10.56RR118 pKa = 11.84VDD120 pKa = 3.73LTLLSLLLVVLMILASIWISMGYY143 pKa = 9.65VYY145 pKa = 11.22YY146 pKa = 11.01SMRR149 pKa = 11.84RR150 pKa = 11.84VSYY153 pKa = 9.82KK154 pKa = 10.19VRR156 pKa = 11.84GIQYY160 pKa = 9.59EE161 pKa = 3.97AMQAGSEE168 pKa = 4.11FRR170 pKa = 11.84EE171 pKa = 4.49GEE173 pKa = 4.08VPKK176 pKa = 10.65FQVRR180 pKa = 11.84VLLPGVFVNSHH191 pKa = 4.53QGYY194 pKa = 9.5GIRR197 pKa = 11.84VEE199 pKa = 3.97NWLVLPTHH207 pKa = 6.38VLTGSEE213 pKa = 3.84ILLEE217 pKa = 4.28GSRR220 pKa = 11.84GRR222 pKa = 11.84VVIPTDD228 pKa = 3.52HH229 pKa = 6.92EE230 pKa = 4.27PSKK233 pKa = 10.43LHH235 pKa = 7.04PDD237 pKa = 3.25VAYY240 pKa = 10.76VWLEE244 pKa = 3.67DD245 pKa = 4.13SQWTTLGTRR254 pKa = 11.84SAKK257 pKa = 9.62TIPITSCHH265 pKa = 5.28VRR267 pKa = 11.84CTGQKK272 pKa = 8.49GYY274 pKa = 11.99SMGTVNKK281 pKa = 9.18TNIIGVLRR289 pKa = 11.84YY290 pKa = 9.93SGSTVPGMSGAAYY303 pKa = 8.9VAQDD307 pKa = 3.55RR308 pKa = 11.84VVGMHH313 pKa = 6.78TGASMEE319 pKa = 4.28KK320 pKa = 9.88NLGVAAALFVNEE332 pKa = 4.62LKK334 pKa = 10.9KK335 pKa = 10.69KK336 pKa = 9.83IKK338 pKa = 10.6NEE340 pKa = 4.28AIFLPSSNNSEE351 pKa = 3.93DD352 pKa = 3.51AAYY355 pKa = 10.69QMKK358 pKa = 10.44SFKK361 pKa = 8.53NTKK364 pKa = 8.7WDD366 pKa = 3.57AKK368 pKa = 10.57LLEE371 pKa = 4.55ALAEE375 pKa = 4.32NADD378 pKa = 4.18DD379 pKa = 4.28MDD381 pKa = 4.65LGSWGGNVDD390 pKa = 6.25FDD392 pKa = 5.12PDD394 pKa = 3.4WKK396 pKa = 11.08AEE398 pKa = 4.13SAEE401 pKa = 4.19EE402 pKa = 3.93PKK404 pKa = 10.51KK405 pKa = 10.76QNPIEE410 pKa = 4.14LATLIMKK417 pKa = 8.52QHH419 pKa = 6.05GPSGEE424 pKa = 4.02EE425 pKa = 3.7TGFSLLNTKK434 pKa = 9.01VLEE437 pKa = 4.94DD438 pKa = 3.39IKK440 pKa = 11.06DD441 pKa = 3.59LQEE444 pKa = 3.88RR445 pKa = 11.84LTIVEE450 pKa = 4.69EE451 pKa = 4.29YY452 pKa = 10.38IKK454 pKa = 10.21TKK456 pKa = 10.11ISKK459 pKa = 10.1KK460 pKa = 10.14KK461 pKa = 8.11EE462 pKa = 3.68PKK464 pKa = 10.18PIVTYY469 pKa = 10.57DD470 pKa = 3.63CEE472 pKa = 4.11EE473 pKa = 4.69CGAKK477 pKa = 7.53TTSEE481 pKa = 4.05SDD483 pKa = 3.51LTKK486 pKa = 10.67HH487 pKa = 4.98QAKK490 pKa = 9.66HH491 pKa = 5.52VKK493 pKa = 9.26HH494 pKa = 6.24KK495 pKa = 11.27CEE497 pKa = 3.96ICDD500 pKa = 3.45ITCQTALRR508 pKa = 11.84LANHH512 pKa = 6.29VANAHH517 pKa = 5.43RR518 pKa = 11.84TAKK521 pKa = 10.39PEE523 pKa = 3.76SAYY526 pKa = 10.17PGDD529 pKa = 3.52ARR531 pKa = 11.84VVVQTAPEE539 pKa = 4.12TQAFLDD545 pKa = 4.0PRR547 pKa = 11.84SNSPKK552 pKa = 9.9RR553 pKa = 11.84KK554 pKa = 8.52NRR556 pKa = 11.84SSKK559 pKa = 10.23RR560 pKa = 11.84SSSTSSKK567 pKa = 10.59KK568 pKa = 9.86QDD570 pKa = 3.33SLPWEE575 pKa = 4.07KK576 pKa = 10.72LLYY579 pKa = 10.78QMVQSQNRR587 pKa = 11.84LEE589 pKa = 4.15EE590 pKa = 4.07YY591 pKa = 10.42LRR593 pKa = 11.84SEE595 pKa = 4.53RR596 pKa = 11.84RR597 pKa = 11.84AMAGPASATQPKK609 pKa = 9.85
Molecular weight: 67.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEK5|A0A1L3KEK5_9VIRU C2H2-type domain-containing protein OS=Hubei diptera virus 13 OX=1922874 PE=4 SV=1
MM1 pKa = 7.33VKK3 pKa = 10.12GKK5 pKa = 10.55SVVCPQCGRR14 pKa = 11.84RR15 pKa = 11.84FASQKK20 pKa = 10.83ALAQHH25 pKa = 6.87RR26 pKa = 11.84LAAHH30 pKa = 6.65AQPAPALRR38 pKa = 11.84RR39 pKa = 11.84AAKK42 pKa = 9.69RR43 pKa = 11.84VVRR46 pKa = 11.84SKK48 pKa = 10.84RR49 pKa = 11.84AGSSLSVQTVNGNDD63 pKa = 4.04RR64 pKa = 11.84IGHH67 pKa = 4.85WEE69 pKa = 3.81IPNNAGSGSLFSVIPIAPYY88 pKa = 9.29MLNDD92 pKa = 3.47TRR94 pKa = 11.84LQAHH98 pKa = 5.9SLLWARR104 pKa = 11.84WRR106 pKa = 11.84PVALKK111 pKa = 10.73VRR113 pKa = 11.84VSLAGASTTFGSVCVGWSPEE133 pKa = 3.89PDD135 pKa = 3.28FSKK138 pKa = 11.01ASSNSLNAQRR148 pKa = 11.84VLSLKK153 pKa = 9.89PSKK156 pKa = 9.66EE157 pKa = 3.53VRR159 pKa = 11.84AWEE162 pKa = 4.1TVEE165 pKa = 4.01MVLPVATDD173 pKa = 3.61RR174 pKa = 11.84KK175 pKa = 8.38WYY177 pKa = 7.75HH178 pKa = 4.68TTGMHH183 pKa = 5.92QEE185 pKa = 4.05ASHH188 pKa = 6.38GCLVVTIDD196 pKa = 4.0SAFGGYY202 pKa = 7.53TGKK205 pKa = 10.25IGFTAHH211 pKa = 7.27LMWTVQFEE219 pKa = 4.4GVEE222 pKa = 4.16LDD224 pKa = 3.89SVAGPVSKK232 pKa = 9.95DD233 pKa = 3.24TIQADD238 pKa = 3.98SGWSHH243 pKa = 7.3FFTTSDD249 pKa = 3.7SGWDD253 pKa = 3.36ATRR256 pKa = 11.84LTFKK260 pKa = 10.26EE261 pKa = 4.16SSGGSMVPFSSAKK274 pKa = 9.99PNFIYY279 pKa = 10.75EE280 pKa = 4.42PDD282 pKa = 3.25SSTRR286 pKa = 11.84VHH288 pKa = 6.21YY289 pKa = 10.67VRR291 pKa = 11.84EE292 pKa = 4.26DD293 pKa = 3.32GKK295 pKa = 11.09EE296 pKa = 3.5ADD298 pKa = 2.89AKK300 pKa = 10.56YY301 pKa = 9.88FVRR304 pKa = 11.84IQDD307 pKa = 3.35YY308 pKa = 7.52STPGLALCASIADD321 pKa = 3.58AKK323 pKa = 11.08AYY325 pKa = 9.54IQTGDD330 pKa = 3.44TTKK333 pKa = 11.09LLPYY337 pKa = 9.62KK338 pKa = 10.34SASSWVTPDD347 pKa = 3.3DD348 pKa = 3.86PKK350 pKa = 11.21FVGKK354 pKa = 9.07PAGSYY359 pKa = 8.11TAVRR363 pKa = 11.84PEE365 pKa = 3.95EE366 pKa = 3.77QDD368 pKa = 3.34LQSEE372 pKa = 4.59IEE374 pKa = 4.14RR375 pKa = 11.84LRR377 pKa = 11.84QQLQLLEE384 pKa = 4.54AGNEE388 pKa = 3.7KK389 pKa = 10.27WEE391 pKa = 4.23VLGSPP396 pKa = 4.47
MM1 pKa = 7.33VKK3 pKa = 10.12GKK5 pKa = 10.55SVVCPQCGRR14 pKa = 11.84RR15 pKa = 11.84FASQKK20 pKa = 10.83ALAQHH25 pKa = 6.87RR26 pKa = 11.84LAAHH30 pKa = 6.65AQPAPALRR38 pKa = 11.84RR39 pKa = 11.84AAKK42 pKa = 9.69RR43 pKa = 11.84VVRR46 pKa = 11.84SKK48 pKa = 10.84RR49 pKa = 11.84AGSSLSVQTVNGNDD63 pKa = 4.04RR64 pKa = 11.84IGHH67 pKa = 4.85WEE69 pKa = 3.81IPNNAGSGSLFSVIPIAPYY88 pKa = 9.29MLNDD92 pKa = 3.47TRR94 pKa = 11.84LQAHH98 pKa = 5.9SLLWARR104 pKa = 11.84WRR106 pKa = 11.84PVALKK111 pKa = 10.73VRR113 pKa = 11.84VSLAGASTTFGSVCVGWSPEE133 pKa = 3.89PDD135 pKa = 3.28FSKK138 pKa = 11.01ASSNSLNAQRR148 pKa = 11.84VLSLKK153 pKa = 9.89PSKK156 pKa = 9.66EE157 pKa = 3.53VRR159 pKa = 11.84AWEE162 pKa = 4.1TVEE165 pKa = 4.01MVLPVATDD173 pKa = 3.61RR174 pKa = 11.84KK175 pKa = 8.38WYY177 pKa = 7.75HH178 pKa = 4.68TTGMHH183 pKa = 5.92QEE185 pKa = 4.05ASHH188 pKa = 6.38GCLVVTIDD196 pKa = 4.0SAFGGYY202 pKa = 7.53TGKK205 pKa = 10.25IGFTAHH211 pKa = 7.27LMWTVQFEE219 pKa = 4.4GVEE222 pKa = 4.16LDD224 pKa = 3.89SVAGPVSKK232 pKa = 9.95DD233 pKa = 3.24TIQADD238 pKa = 3.98SGWSHH243 pKa = 7.3FFTTSDD249 pKa = 3.7SGWDD253 pKa = 3.36ATRR256 pKa = 11.84LTFKK260 pKa = 10.26EE261 pKa = 4.16SSGGSMVPFSSAKK274 pKa = 9.99PNFIYY279 pKa = 10.75EE280 pKa = 4.42PDD282 pKa = 3.25SSTRR286 pKa = 11.84VHH288 pKa = 6.21YY289 pKa = 10.67VRR291 pKa = 11.84EE292 pKa = 4.26DD293 pKa = 3.32GKK295 pKa = 11.09EE296 pKa = 3.5ADD298 pKa = 2.89AKK300 pKa = 10.56YY301 pKa = 9.88FVRR304 pKa = 11.84IQDD307 pKa = 3.35YY308 pKa = 7.52STPGLALCASIADD321 pKa = 3.58AKK323 pKa = 11.08AYY325 pKa = 9.54IQTGDD330 pKa = 3.44TTKK333 pKa = 11.09LLPYY337 pKa = 9.62KK338 pKa = 10.34SASSWVTPDD347 pKa = 3.3DD348 pKa = 3.86PKK350 pKa = 11.21FVGKK354 pKa = 9.07PAGSYY359 pKa = 8.11TAVRR363 pKa = 11.84PEE365 pKa = 3.95EE366 pKa = 3.77QDD368 pKa = 3.34LQSEE372 pKa = 4.59IEE374 pKa = 4.14RR375 pKa = 11.84LRR377 pKa = 11.84QQLQLLEE384 pKa = 4.54AGNEE388 pKa = 3.7KK389 pKa = 10.27WEE391 pKa = 4.23VLGSPP396 pKa = 4.47
Molecular weight: 43.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1443 |
396 |
609 |
481.0 |
53.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.524 ± 0.623 | 1.455 ± 0.1 |
5.128 ± 0.512 | 6.306 ± 0.589 |
3.049 ± 0.485 | 6.514 ± 0.348 |
2.356 ± 0.116 | 4.019 ± 0.393 |
6.999 ± 0.52 | 8.801 ± 0.574 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.772 ± 0.582 | 2.772 ± 0.271 |
4.92 ± 0.278 | 3.534 ± 0.418 |
5.821 ± 0.651 | 8.108 ± 1.319 |
5.336 ± 0.666 | 8.039 ± 0.215 |
2.356 ± 0.376 | 3.188 ± 0.254 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
