
Capybara microvirus Cap1_SP_93
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.37
Get precalculated fractions of proteins
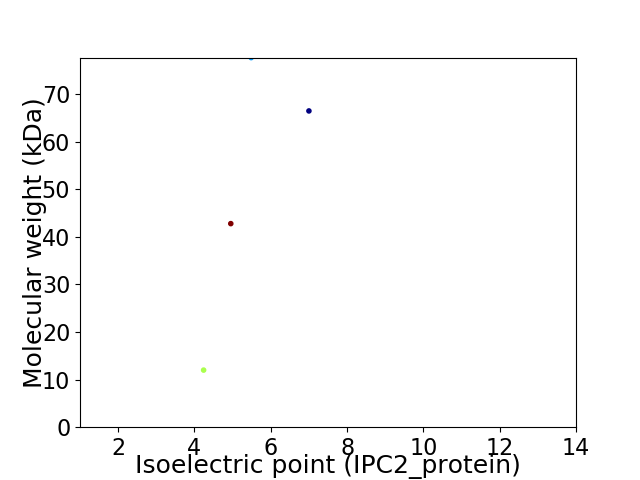
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
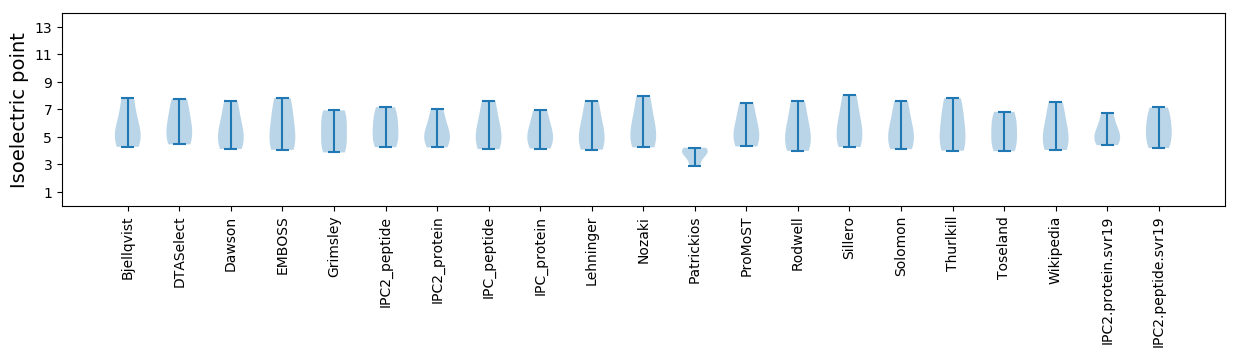
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W424|A0A4P8W424_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_93 OX=2584799 PE=4 SV=1
MM1 pKa = 7.72AFFPLNFFGASICEE15 pKa = 3.88FVPTEE20 pKa = 3.76KK21 pKa = 10.33RR22 pKa = 11.84YY23 pKa = 9.94ILSKK27 pKa = 10.99SIDD30 pKa = 3.68PNNEE34 pKa = 3.67NNVLTSGVLDD44 pKa = 3.91DD45 pKa = 5.12VPIFEE50 pKa = 4.93NSKK53 pKa = 10.63DD54 pKa = 4.0LPNPDD59 pKa = 3.76DD60 pKa = 3.57TDD62 pKa = 3.71LARR65 pKa = 11.84QLLLGSPIQDD75 pKa = 3.29YY76 pKa = 11.39SGTKK80 pKa = 9.26ILNGKK85 pKa = 8.12EE86 pKa = 3.98LVISDD91 pKa = 4.04SQANDD96 pKa = 3.22YY97 pKa = 10.61LAEE100 pKa = 4.18LEE102 pKa = 4.21KK103 pKa = 10.57TLVKK107 pKa = 10.82NN108 pKa = 3.91
MM1 pKa = 7.72AFFPLNFFGASICEE15 pKa = 3.88FVPTEE20 pKa = 3.76KK21 pKa = 10.33RR22 pKa = 11.84YY23 pKa = 9.94ILSKK27 pKa = 10.99SIDD30 pKa = 3.68PNNEE34 pKa = 3.67NNVLTSGVLDD44 pKa = 3.91DD45 pKa = 5.12VPIFEE50 pKa = 4.93NSKK53 pKa = 10.63DD54 pKa = 4.0LPNPDD59 pKa = 3.76DD60 pKa = 3.57TDD62 pKa = 3.71LARR65 pKa = 11.84QLLLGSPIQDD75 pKa = 3.29YY76 pKa = 11.39SGTKK80 pKa = 9.26ILNGKK85 pKa = 8.12EE86 pKa = 3.98LVISDD91 pKa = 4.04SQANDD96 pKa = 3.22YY97 pKa = 10.61LAEE100 pKa = 4.18LEE102 pKa = 4.21KK103 pKa = 10.57TLVKK107 pKa = 10.82NN108 pKa = 3.91
Molecular weight: 12.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVN5|A0A4V1FVN5_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_93 OX=2584799 PE=4 SV=1
MM1 pKa = 7.49YY2 pKa = 11.11NMSTCYY8 pKa = 10.73NKK10 pKa = 9.97IEE12 pKa = 4.13IKK14 pKa = 10.62NPSVHH19 pKa = 5.89FRR21 pKa = 11.84KK22 pKa = 10.51NMDD25 pKa = 3.52KK26 pKa = 11.09LLLQVDD32 pKa = 4.65CGRR35 pKa = 11.84CDD37 pKa = 2.92SCLSQKK43 pKa = 10.39RR44 pKa = 11.84NNWYY48 pKa = 9.36VRR50 pKa = 11.84ARR52 pKa = 11.84FEE54 pKa = 5.31LMACLGQDD62 pKa = 3.0KK63 pKa = 10.17DD64 pKa = 3.64GHH66 pKa = 5.66YY67 pKa = 9.99VAPSGFCLFPTLTYY81 pKa = 10.69LSYY84 pKa = 10.85KK85 pKa = 10.18RR86 pKa = 11.84PSLHH90 pKa = 7.37IIDD93 pKa = 4.91EE94 pKa = 4.86FGDD97 pKa = 3.35DD98 pKa = 3.35WFFEE102 pKa = 4.45GFSYY106 pKa = 9.9QHH108 pKa = 5.62ITDD111 pKa = 3.95YY112 pKa = 11.65VNTIQTQYY120 pKa = 9.22KK121 pKa = 9.52RR122 pKa = 11.84KK123 pKa = 9.71YY124 pKa = 8.88NKK126 pKa = 9.27PDD128 pKa = 3.49DD129 pKa = 3.9GVRR132 pKa = 11.84YY133 pKa = 7.32MVCSEE138 pKa = 3.97YY139 pKa = 11.12GGNAEE144 pKa = 4.15YY145 pKa = 10.7LDD147 pKa = 4.0EE148 pKa = 5.7RR149 pKa = 11.84GLPRR153 pKa = 11.84IAQAAPHH160 pKa = 4.89YY161 pKa = 9.89HH162 pKa = 6.65CLLFFPARR170 pKa = 11.84ASKK173 pKa = 10.9DD174 pKa = 3.21SLYY177 pKa = 10.49WKK179 pKa = 10.19DD180 pKa = 3.56YY181 pKa = 10.96VSNLWRR187 pKa = 11.84KK188 pKa = 7.57QHH190 pKa = 6.92DD191 pKa = 3.87NALVGYY197 pKa = 8.85SKK199 pKa = 11.16SKK201 pKa = 9.96GAFVRR206 pKa = 11.84DD207 pKa = 3.51CDD209 pKa = 4.15AIKK212 pKa = 10.81YY213 pKa = 6.01VTSYY217 pKa = 10.5CHH219 pKa = 6.37KK220 pKa = 10.64QFDD223 pKa = 4.68YY224 pKa = 10.73YY225 pKa = 10.94QRR227 pKa = 11.84PEE229 pKa = 3.6LDD231 pKa = 2.83AYY233 pKa = 9.63IWHH236 pKa = 7.84DD237 pKa = 3.84YY238 pKa = 10.56IKK240 pKa = 10.87DD241 pKa = 3.48GFRR244 pKa = 11.84FQIPPMSQWKK254 pKa = 10.32DD255 pKa = 3.55NEE257 pKa = 3.92KK258 pKa = 10.99LEE260 pKa = 4.0YY261 pKa = 11.01LEE263 pKa = 4.03ILKK266 pKa = 10.4SRR268 pKa = 11.84RR269 pKa = 11.84EE270 pKa = 3.98EE271 pKa = 4.06LKK273 pKa = 10.9SIMPNHH279 pKa = 6.87WEE281 pKa = 3.91SGHH284 pKa = 6.55FGEE287 pKa = 6.22NCAKK291 pKa = 9.72WLSNLSLDD299 pKa = 3.48EE300 pKa = 4.11RR301 pKa = 11.84VNILSNGITFANDD314 pKa = 2.86TNLEE318 pKa = 4.04GKK320 pKa = 9.71LNHH323 pKa = 6.02YY324 pKa = 8.04VVPKK328 pKa = 10.54YY329 pKa = 10.62IFDD332 pKa = 3.67KK333 pKa = 10.8LYY335 pKa = 11.15YY336 pKa = 9.76DD337 pKa = 3.29IQYY340 pKa = 9.94QRR342 pKa = 11.84VYY344 pKa = 11.28DD345 pKa = 3.93EE346 pKa = 5.17DD347 pKa = 4.57LDD349 pKa = 4.93RR350 pKa = 11.84EE351 pKa = 4.48VLRR354 pKa = 11.84PALKK358 pKa = 10.4KK359 pKa = 9.54PTDD362 pKa = 3.65ILCKK366 pKa = 10.6DD367 pKa = 3.97FVSLFRR373 pKa = 11.84KK374 pKa = 9.81VYY376 pKa = 10.17NDD378 pKa = 3.28KK379 pKa = 10.82LLKK382 pKa = 10.19FNHH385 pKa = 6.53LFRR388 pKa = 11.84YY389 pKa = 7.92ITLCDD394 pKa = 3.51LTDD397 pKa = 3.9NEE399 pKa = 4.46ILQLKK404 pKa = 10.07NIDD407 pKa = 3.44NKK409 pKa = 10.59RR410 pKa = 11.84DD411 pKa = 3.62YY412 pKa = 10.58VKK414 pKa = 10.73NWTRR418 pKa = 11.84EE419 pKa = 3.77DD420 pKa = 3.68LYY422 pKa = 11.41SYY424 pKa = 11.18FEE426 pKa = 4.73DD427 pKa = 4.11FQSRR431 pKa = 11.84FSMEE435 pKa = 3.85RR436 pKa = 11.84LVSYY440 pKa = 10.37SYY442 pKa = 10.92IRR444 pKa = 11.84NTLFDD449 pKa = 4.51INTSLDD455 pKa = 3.85LLQMSDD461 pKa = 3.75EE462 pKa = 4.45DD463 pKa = 4.4FISNVEE469 pKa = 3.74DD470 pKa = 3.19HH471 pKa = 7.08KK472 pKa = 11.29IYY474 pKa = 11.03VSLSNKK480 pKa = 9.8SFSLKK485 pKa = 10.26KK486 pKa = 10.38GAEE489 pKa = 4.17PYY491 pKa = 10.43YY492 pKa = 10.94KK493 pKa = 10.42SDD495 pKa = 3.91DD496 pKa = 3.7YY497 pKa = 11.72KK498 pKa = 11.45HH499 pKa = 6.7NIRR502 pKa = 11.84NLIPYY507 pKa = 9.52RR508 pKa = 11.84EE509 pKa = 4.13PEE511 pKa = 3.86KK512 pKa = 11.04EE513 pKa = 4.21FILEE517 pKa = 3.83LGEE520 pKa = 4.42RR521 pKa = 11.84INLIKK526 pKa = 10.76SKK528 pKa = 10.72NRR530 pKa = 11.84LNAYY534 pKa = 7.42HH535 pKa = 7.57TIRR538 pKa = 11.84KK539 pKa = 8.13LQQQDD544 pKa = 3.5KK545 pKa = 10.18QNHH548 pKa = 6.51LLTSLIVV555 pKa = 3.26
MM1 pKa = 7.49YY2 pKa = 11.11NMSTCYY8 pKa = 10.73NKK10 pKa = 9.97IEE12 pKa = 4.13IKK14 pKa = 10.62NPSVHH19 pKa = 5.89FRR21 pKa = 11.84KK22 pKa = 10.51NMDD25 pKa = 3.52KK26 pKa = 11.09LLLQVDD32 pKa = 4.65CGRR35 pKa = 11.84CDD37 pKa = 2.92SCLSQKK43 pKa = 10.39RR44 pKa = 11.84NNWYY48 pKa = 9.36VRR50 pKa = 11.84ARR52 pKa = 11.84FEE54 pKa = 5.31LMACLGQDD62 pKa = 3.0KK63 pKa = 10.17DD64 pKa = 3.64GHH66 pKa = 5.66YY67 pKa = 9.99VAPSGFCLFPTLTYY81 pKa = 10.69LSYY84 pKa = 10.85KK85 pKa = 10.18RR86 pKa = 11.84PSLHH90 pKa = 7.37IIDD93 pKa = 4.91EE94 pKa = 4.86FGDD97 pKa = 3.35DD98 pKa = 3.35WFFEE102 pKa = 4.45GFSYY106 pKa = 9.9QHH108 pKa = 5.62ITDD111 pKa = 3.95YY112 pKa = 11.65VNTIQTQYY120 pKa = 9.22KK121 pKa = 9.52RR122 pKa = 11.84KK123 pKa = 9.71YY124 pKa = 8.88NKK126 pKa = 9.27PDD128 pKa = 3.49DD129 pKa = 3.9GVRR132 pKa = 11.84YY133 pKa = 7.32MVCSEE138 pKa = 3.97YY139 pKa = 11.12GGNAEE144 pKa = 4.15YY145 pKa = 10.7LDD147 pKa = 4.0EE148 pKa = 5.7RR149 pKa = 11.84GLPRR153 pKa = 11.84IAQAAPHH160 pKa = 4.89YY161 pKa = 9.89HH162 pKa = 6.65CLLFFPARR170 pKa = 11.84ASKK173 pKa = 10.9DD174 pKa = 3.21SLYY177 pKa = 10.49WKK179 pKa = 10.19DD180 pKa = 3.56YY181 pKa = 10.96VSNLWRR187 pKa = 11.84KK188 pKa = 7.57QHH190 pKa = 6.92DD191 pKa = 3.87NALVGYY197 pKa = 8.85SKK199 pKa = 11.16SKK201 pKa = 9.96GAFVRR206 pKa = 11.84DD207 pKa = 3.51CDD209 pKa = 4.15AIKK212 pKa = 10.81YY213 pKa = 6.01VTSYY217 pKa = 10.5CHH219 pKa = 6.37KK220 pKa = 10.64QFDD223 pKa = 4.68YY224 pKa = 10.73YY225 pKa = 10.94QRR227 pKa = 11.84PEE229 pKa = 3.6LDD231 pKa = 2.83AYY233 pKa = 9.63IWHH236 pKa = 7.84DD237 pKa = 3.84YY238 pKa = 10.56IKK240 pKa = 10.87DD241 pKa = 3.48GFRR244 pKa = 11.84FQIPPMSQWKK254 pKa = 10.32DD255 pKa = 3.55NEE257 pKa = 3.92KK258 pKa = 10.99LEE260 pKa = 4.0YY261 pKa = 11.01LEE263 pKa = 4.03ILKK266 pKa = 10.4SRR268 pKa = 11.84RR269 pKa = 11.84EE270 pKa = 3.98EE271 pKa = 4.06LKK273 pKa = 10.9SIMPNHH279 pKa = 6.87WEE281 pKa = 3.91SGHH284 pKa = 6.55FGEE287 pKa = 6.22NCAKK291 pKa = 9.72WLSNLSLDD299 pKa = 3.48EE300 pKa = 4.11RR301 pKa = 11.84VNILSNGITFANDD314 pKa = 2.86TNLEE318 pKa = 4.04GKK320 pKa = 9.71LNHH323 pKa = 6.02YY324 pKa = 8.04VVPKK328 pKa = 10.54YY329 pKa = 10.62IFDD332 pKa = 3.67KK333 pKa = 10.8LYY335 pKa = 11.15YY336 pKa = 9.76DD337 pKa = 3.29IQYY340 pKa = 9.94QRR342 pKa = 11.84VYY344 pKa = 11.28DD345 pKa = 3.93EE346 pKa = 5.17DD347 pKa = 4.57LDD349 pKa = 4.93RR350 pKa = 11.84EE351 pKa = 4.48VLRR354 pKa = 11.84PALKK358 pKa = 10.4KK359 pKa = 9.54PTDD362 pKa = 3.65ILCKK366 pKa = 10.6DD367 pKa = 3.97FVSLFRR373 pKa = 11.84KK374 pKa = 9.81VYY376 pKa = 10.17NDD378 pKa = 3.28KK379 pKa = 10.82LLKK382 pKa = 10.19FNHH385 pKa = 6.53LFRR388 pKa = 11.84YY389 pKa = 7.92ITLCDD394 pKa = 3.51LTDD397 pKa = 3.9NEE399 pKa = 4.46ILQLKK404 pKa = 10.07NIDD407 pKa = 3.44NKK409 pKa = 10.59RR410 pKa = 11.84DD411 pKa = 3.62YY412 pKa = 10.58VKK414 pKa = 10.73NWTRR418 pKa = 11.84EE419 pKa = 3.77DD420 pKa = 3.68LYY422 pKa = 11.41SYY424 pKa = 11.18FEE426 pKa = 4.73DD427 pKa = 4.11FQSRR431 pKa = 11.84FSMEE435 pKa = 3.85RR436 pKa = 11.84LVSYY440 pKa = 10.37SYY442 pKa = 10.92IRR444 pKa = 11.84NTLFDD449 pKa = 4.51INTSLDD455 pKa = 3.85LLQMSDD461 pKa = 3.75EE462 pKa = 4.45DD463 pKa = 4.4FISNVEE469 pKa = 3.74DD470 pKa = 3.19HH471 pKa = 7.08KK472 pKa = 11.29IYY474 pKa = 11.03VSLSNKK480 pKa = 9.8SFSLKK485 pKa = 10.26KK486 pKa = 10.38GAEE489 pKa = 4.17PYY491 pKa = 10.43YY492 pKa = 10.94KK493 pKa = 10.42SDD495 pKa = 3.91DD496 pKa = 3.7YY497 pKa = 11.72KK498 pKa = 11.45HH499 pKa = 6.7NIRR502 pKa = 11.84NLIPYY507 pKa = 9.52RR508 pKa = 11.84EE509 pKa = 4.13PEE511 pKa = 3.86KK512 pKa = 11.04EE513 pKa = 4.21FILEE517 pKa = 3.83LGEE520 pKa = 4.42RR521 pKa = 11.84INLIKK526 pKa = 10.76SKK528 pKa = 10.72NRR530 pKa = 11.84LNAYY534 pKa = 7.42HH535 pKa = 7.57TIRR538 pKa = 11.84KK539 pKa = 8.13LQQQDD544 pKa = 3.5KK545 pKa = 10.18QNHH548 pKa = 6.51LLTSLIVV555 pKa = 3.26
Molecular weight: 66.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1728 |
108 |
678 |
432.0 |
49.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.382 ± 1.37 | 1.505 ± 0.412 |
7.465 ± 0.481 | 4.514 ± 0.898 |
5.035 ± 0.954 | 5.208 ± 0.714 |
2.199 ± 0.691 | 5.845 ± 0.102 |
6.076 ± 1.015 | 8.507 ± 1.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.652 | 6.655 ± 0.379 |
3.993 ± 0.999 | 5.324 ± 1.201 |
4.282 ± 0.708 | 8.044 ± 0.674 |
5.324 ± 1.199 | 4.745 ± 0.252 |
1.62 ± 0.29 | 5.787 ± 0.976 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
