
Prevotella copri CAG:164
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Prevotellaceae; Prevotella; environmental samples
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
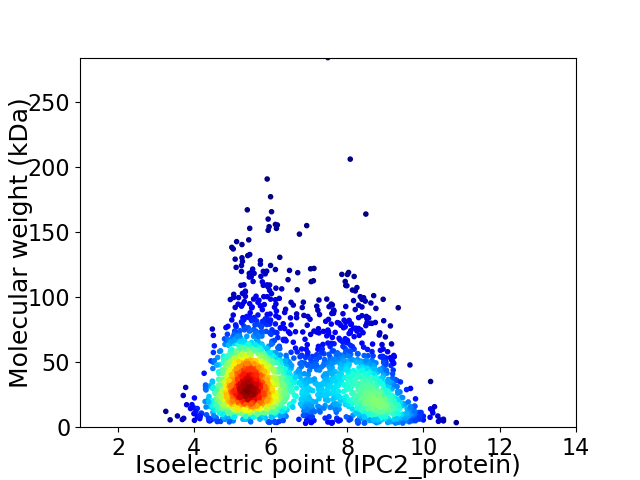
Virtual 2D-PAGE plot for 2331 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6BQZ2|R6BQZ2_9BACT Putative lipoprotein OS=Prevotella copri CAG:164 OX=1263102 GN=BN510_00163 PE=4 SV=1
MM1 pKa = 7.29EE2 pKa = 4.34QNKK5 pKa = 10.08NNGTTSAAKK14 pKa = 9.26WKK16 pKa = 9.42EE17 pKa = 3.72VLIGGVSGIALGTAGTLFASSIPLEE42 pKa = 4.5ASAEE46 pKa = 4.25TPEE49 pKa = 4.36TPEE52 pKa = 4.82SEE54 pKa = 4.29DD55 pKa = 3.65ANVGNGGEE63 pKa = 4.14NVTAQMATCVTDD75 pKa = 3.53SMSFSAAFGAARR87 pKa = 11.84AEE89 pKa = 4.18VGPGGVFEE97 pKa = 4.22WHH99 pKa = 6.36GNLYY103 pKa = 8.62GTYY106 pKa = 9.4TADD109 pKa = 3.14EE110 pKa = 4.56WNSMDD115 pKa = 3.34SDD117 pKa = 3.49ARR119 pKa = 11.84NEE121 pKa = 4.14YY122 pKa = 10.77AEE124 pKa = 4.59SIHH127 pKa = 6.58WDD129 pKa = 4.11GPSHH133 pKa = 6.75EE134 pKa = 4.68YY135 pKa = 10.29VAASTSHH142 pKa = 6.28SAHH145 pKa = 6.73TYY147 pKa = 6.25TANHH151 pKa = 5.9SVADD155 pKa = 4.17SVCQQQTPPEE165 pKa = 4.5KK166 pKa = 10.3DD167 pKa = 3.58DD168 pKa = 4.34EE169 pKa = 4.34EE170 pKa = 4.73TPDD173 pKa = 3.43VEE175 pKa = 4.91IIGVEE180 pKa = 3.8HH181 pKa = 6.26TNIDD185 pKa = 3.78GEE187 pKa = 4.21HH188 pKa = 7.04DD189 pKa = 4.37SIIGSASVNGQAVYY203 pKa = 9.95FIDD206 pKa = 5.25VDD208 pKa = 3.82GQDD211 pKa = 3.72NEE213 pKa = 4.42FEE215 pKa = 5.22LMVTDD220 pKa = 5.18ANGNQQIDD228 pKa = 3.65EE229 pKa = 4.67GEE231 pKa = 4.23VVDD234 pKa = 6.41ISDD237 pKa = 3.17QHH239 pKa = 5.88MSVSHH244 pKa = 6.08FEE246 pKa = 3.99EE247 pKa = 4.38MAQANHH253 pKa = 6.7TSDD256 pKa = 3.83TDD258 pKa = 3.71NDD260 pKa = 4.0DD261 pKa = 3.85TPVQYY266 pKa = 10.67YY267 pKa = 11.04ANNEE271 pKa = 4.09DD272 pKa = 4.02LPDD275 pKa = 3.86YY276 pKa = 11.6VNDD279 pKa = 4.46ADD281 pKa = 4.87PGTLAA286 pKa = 4.79
MM1 pKa = 7.29EE2 pKa = 4.34QNKK5 pKa = 10.08NNGTTSAAKK14 pKa = 9.26WKK16 pKa = 9.42EE17 pKa = 3.72VLIGGVSGIALGTAGTLFASSIPLEE42 pKa = 4.5ASAEE46 pKa = 4.25TPEE49 pKa = 4.36TPEE52 pKa = 4.82SEE54 pKa = 4.29DD55 pKa = 3.65ANVGNGGEE63 pKa = 4.14NVTAQMATCVTDD75 pKa = 3.53SMSFSAAFGAARR87 pKa = 11.84AEE89 pKa = 4.18VGPGGVFEE97 pKa = 4.22WHH99 pKa = 6.36GNLYY103 pKa = 8.62GTYY106 pKa = 9.4TADD109 pKa = 3.14EE110 pKa = 4.56WNSMDD115 pKa = 3.34SDD117 pKa = 3.49ARR119 pKa = 11.84NEE121 pKa = 4.14YY122 pKa = 10.77AEE124 pKa = 4.59SIHH127 pKa = 6.58WDD129 pKa = 4.11GPSHH133 pKa = 6.75EE134 pKa = 4.68YY135 pKa = 10.29VAASTSHH142 pKa = 6.28SAHH145 pKa = 6.73TYY147 pKa = 6.25TANHH151 pKa = 5.9SVADD155 pKa = 4.17SVCQQQTPPEE165 pKa = 4.5KK166 pKa = 10.3DD167 pKa = 3.58DD168 pKa = 4.34EE169 pKa = 4.34EE170 pKa = 4.73TPDD173 pKa = 3.43VEE175 pKa = 4.91IIGVEE180 pKa = 3.8HH181 pKa = 6.26TNIDD185 pKa = 3.78GEE187 pKa = 4.21HH188 pKa = 7.04DD189 pKa = 4.37SIIGSASVNGQAVYY203 pKa = 9.95FIDD206 pKa = 5.25VDD208 pKa = 3.82GQDD211 pKa = 3.72NEE213 pKa = 4.42FEE215 pKa = 5.22LMVTDD220 pKa = 5.18ANGNQQIDD228 pKa = 3.65EE229 pKa = 4.67GEE231 pKa = 4.23VVDD234 pKa = 6.41ISDD237 pKa = 3.17QHH239 pKa = 5.88MSVSHH244 pKa = 6.08FEE246 pKa = 3.99EE247 pKa = 4.38MAQANHH253 pKa = 6.7TSDD256 pKa = 3.83TDD258 pKa = 3.71NDD260 pKa = 4.0DD261 pKa = 3.85TPVQYY266 pKa = 10.67YY267 pKa = 11.04ANNEE271 pKa = 4.09DD272 pKa = 4.02LPDD275 pKa = 3.86YY276 pKa = 11.6VNDD279 pKa = 4.46ADD281 pKa = 4.87PGTLAA286 pKa = 4.79
Molecular weight: 30.52 kDa
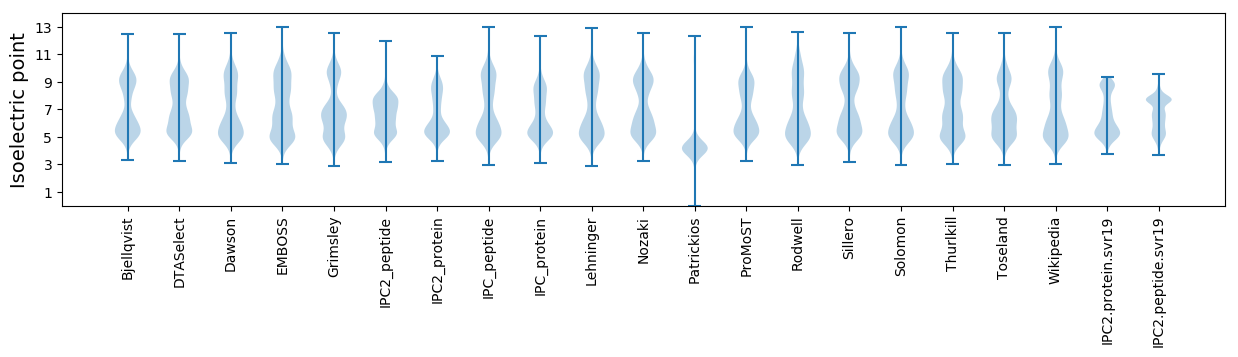
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6BPP6|R6BPP6_9BACT ATP phosphoribosyltransferase OS=Prevotella copri CAG:164 OX=1263102 GN=hisG PE=3 SV=1
MM1 pKa = 7.85PNGKK5 pKa = 9.19KK6 pKa = 10.25KK7 pKa = 10.12KK8 pKa = 7.0GHH10 pKa = 6.14KK11 pKa = 9.06MATHH15 pKa = 6.13KK16 pKa = 10.39RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.25NRR25 pKa = 11.84HH26 pKa = 4.69KK27 pKa = 11.1SKK29 pKa = 11.1
MM1 pKa = 7.85PNGKK5 pKa = 9.19KK6 pKa = 10.25KK7 pKa = 10.12KK8 pKa = 7.0GHH10 pKa = 6.14KK11 pKa = 9.06MATHH15 pKa = 6.13KK16 pKa = 10.39RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.25NRR25 pKa = 11.84HH26 pKa = 4.69KK27 pKa = 11.1SKK29 pKa = 11.1
Molecular weight: 3.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
805592 |
29 |
2512 |
345.6 |
38.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.467 ± 0.046 | 1.31 ± 0.017 |
5.762 ± 0.034 | 6.432 ± 0.053 |
4.428 ± 0.035 | 6.905 ± 0.052 |
2.121 ± 0.025 | 6.743 ± 0.043 |
7.195 ± 0.042 | 8.779 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.089 ± 0.023 | 5.058 ± 0.037 |
3.501 ± 0.028 | 3.723 ± 0.028 |
4.299 ± 0.036 | 5.834 ± 0.043 |
5.367 ± 0.036 | 6.486 ± 0.039 |
1.257 ± 0.019 | 4.242 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
