
Tamlana nanhaiensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tamlana
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
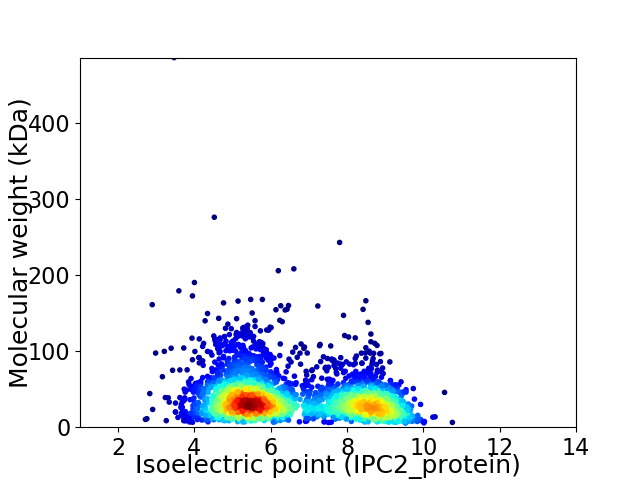
Virtual 2D-PAGE plot for 3029 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D7W1C4|A0A0D7W1C4_9FLAO Mechanosensitive ion channel protein MscS OS=Tamlana nanhaiensis OX=1382798 GN=PK35_07840 PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 10.34RR3 pKa = 11.84LNLAKK8 pKa = 9.77TLALTLIFSIIFMSCEE24 pKa = 3.53NDD26 pKa = 3.83DD27 pKa = 4.32NNTSNEE33 pKa = 4.21TMTSTYY39 pKa = 10.66LYY41 pKa = 10.05TSNNANGNITYY52 pKa = 10.52YY53 pKa = 11.03NVGDD57 pKa = 3.96MSNVMMTTLVTSSTAADD74 pKa = 3.83GVFYY78 pKa = 10.99DD79 pKa = 5.0ADD81 pKa = 4.27DD82 pKa = 4.58DD83 pKa = 4.3MLVQASRR90 pKa = 11.84SNMMLEE96 pKa = 4.19GFMDD100 pKa = 4.0TSMSSTGASISIGISGSSDD119 pKa = 2.73MMSPRR124 pKa = 11.84EE125 pKa = 3.71LAVNGDD131 pKa = 4.23YY132 pKa = 11.25YY133 pKa = 11.62VVADD137 pKa = 4.07NADD140 pKa = 3.22VDD142 pKa = 4.34GDD144 pKa = 4.31PDD146 pKa = 3.93TPDD149 pKa = 2.95GKK151 pKa = 10.94LYY153 pKa = 11.18VYY155 pKa = 10.01MKK157 pKa = 10.36DD158 pKa = 3.72GNDD161 pKa = 3.05FTLRR165 pKa = 11.84NVITTDD171 pKa = 2.67IKK173 pKa = 10.86LWGITFDD180 pKa = 4.37GEE182 pKa = 4.07SLYY185 pKa = 11.05AIVDD189 pKa = 3.84TTNEE193 pKa = 3.76LAVYY197 pKa = 10.87SNFLANTGDD206 pKa = 3.58MMLSATKK213 pKa = 9.71TIAVEE218 pKa = 4.8GIVRR222 pKa = 11.84THH224 pKa = 6.4GLTYY228 pKa = 10.88DD229 pKa = 3.48AMSDD233 pKa = 3.65TMVLTDD239 pKa = 4.14IAAAANGQDD248 pKa = 3.71DD249 pKa = 4.51GGFHH253 pKa = 7.69IITDD257 pKa = 4.45FMSKK261 pKa = 10.26FNNTADD267 pKa = 3.57GGTLAMSDD275 pKa = 3.91QIRR278 pKa = 11.84VAGSSTMLGNPVDD291 pKa = 3.67VAFDD295 pKa = 3.94SEE297 pKa = 4.83TEE299 pKa = 4.04TVYY302 pKa = 10.57IAEE305 pKa = 4.59AGNNGGRR312 pKa = 11.84ILAFNNIGSGGNIAPVLNYY331 pKa = 10.49DD332 pKa = 3.99LASASSVYY340 pKa = 9.98LVKK343 pKa = 10.92YY344 pKa = 10.24
MM1 pKa = 7.54KK2 pKa = 10.34RR3 pKa = 11.84LNLAKK8 pKa = 9.77TLALTLIFSIIFMSCEE24 pKa = 3.53NDD26 pKa = 3.83DD27 pKa = 4.32NNTSNEE33 pKa = 4.21TMTSTYY39 pKa = 10.66LYY41 pKa = 10.05TSNNANGNITYY52 pKa = 10.52YY53 pKa = 11.03NVGDD57 pKa = 3.96MSNVMMTTLVTSSTAADD74 pKa = 3.83GVFYY78 pKa = 10.99DD79 pKa = 5.0ADD81 pKa = 4.27DD82 pKa = 4.58DD83 pKa = 4.3MLVQASRR90 pKa = 11.84SNMMLEE96 pKa = 4.19GFMDD100 pKa = 4.0TSMSSTGASISIGISGSSDD119 pKa = 2.73MMSPRR124 pKa = 11.84EE125 pKa = 3.71LAVNGDD131 pKa = 4.23YY132 pKa = 11.25YY133 pKa = 11.62VVADD137 pKa = 4.07NADD140 pKa = 3.22VDD142 pKa = 4.34GDD144 pKa = 4.31PDD146 pKa = 3.93TPDD149 pKa = 2.95GKK151 pKa = 10.94LYY153 pKa = 11.18VYY155 pKa = 10.01MKK157 pKa = 10.36DD158 pKa = 3.72GNDD161 pKa = 3.05FTLRR165 pKa = 11.84NVITTDD171 pKa = 2.67IKK173 pKa = 10.86LWGITFDD180 pKa = 4.37GEE182 pKa = 4.07SLYY185 pKa = 11.05AIVDD189 pKa = 3.84TTNEE193 pKa = 3.76LAVYY197 pKa = 10.87SNFLANTGDD206 pKa = 3.58MMLSATKK213 pKa = 9.71TIAVEE218 pKa = 4.8GIVRR222 pKa = 11.84THH224 pKa = 6.4GLTYY228 pKa = 10.88DD229 pKa = 3.48AMSDD233 pKa = 3.65TMVLTDD239 pKa = 4.14IAAAANGQDD248 pKa = 3.71DD249 pKa = 4.51GGFHH253 pKa = 7.69IITDD257 pKa = 4.45FMSKK261 pKa = 10.26FNNTADD267 pKa = 3.57GGTLAMSDD275 pKa = 3.91QIRR278 pKa = 11.84VAGSSTMLGNPVDD291 pKa = 3.67VAFDD295 pKa = 3.94SEE297 pKa = 4.83TEE299 pKa = 4.04TVYY302 pKa = 10.57IAEE305 pKa = 4.59AGNNGGRR312 pKa = 11.84ILAFNNIGSGGNIAPVLNYY331 pKa = 10.49DD332 pKa = 3.99LASASSVYY340 pKa = 9.98LVKK343 pKa = 10.92YY344 pKa = 10.24
Molecular weight: 36.78 kDa
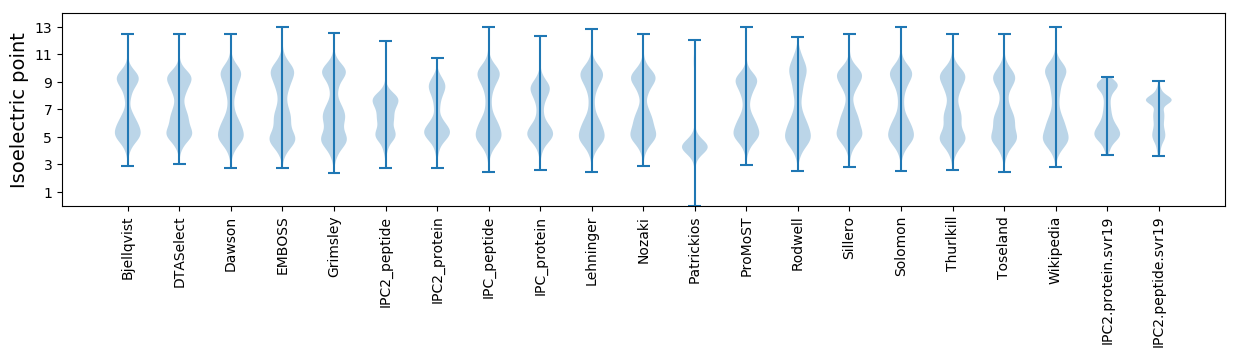
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D7W3V5|A0A0D7W3V5_9FLAO Uncharacterized protein OS=Tamlana nanhaiensis OX=1382798 GN=PK35_03315 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1091831 |
46 |
4626 |
360.5 |
40.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.604 ± 0.043 | 0.754 ± 0.013 |
5.589 ± 0.036 | 6.343 ± 0.035 |
5.275 ± 0.032 | 6.264 ± 0.038 |
1.767 ± 0.022 | 7.721 ± 0.038 |
7.758 ± 0.055 | 9.161 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.025 ± 0.017 | 6.828 ± 0.047 |
3.334 ± 0.02 | 3.331 ± 0.026 |
3.135 ± 0.026 | 6.389 ± 0.036 |
6.107 ± 0.048 | 6.356 ± 0.032 |
1.099 ± 0.018 | 4.16 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
