
Thermoplasmatales archaeon SCGC AB-539-C06
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Thermoplasmatales; unclassified Thermoplasmatales
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
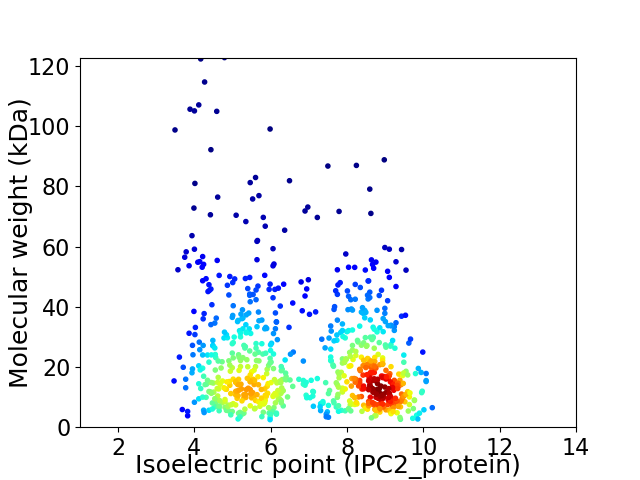
Virtual 2D-PAGE plot for 780 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|N6WMI6|N6WMI6_9ARCH Uncharacterized protein (Fragment) OS=Thermoplasmatales archaeon SCGC AB-539-C06 OX=1242690 GN=MBGDC06_00790 PE=4 SV=1
MM1 pKa = 7.34GKK3 pKa = 10.1KK4 pKa = 10.02SVILLLIGLLIFSNSGFNQIEE25 pKa = 3.98NTKK28 pKa = 10.44AVEE31 pKa = 4.05TTFYY35 pKa = 11.31VDD37 pKa = 5.02DD38 pKa = 6.15DD39 pKa = 4.13GLEE42 pKa = 4.37DD43 pKa = 3.8YY44 pKa = 11.85VNIQYY49 pKa = 10.85AIDD52 pKa = 3.66NASIGDD58 pKa = 3.82TVYY61 pKa = 10.86VYY63 pKa = 10.39HH64 pKa = 7.4GYY66 pKa = 10.4YY67 pKa = 9.92KK68 pKa = 10.84EE69 pKa = 3.8NLVINKK75 pKa = 9.52SISLIGEE82 pKa = 4.17DD83 pKa = 4.84KK84 pKa = 11.46GNTTIDD90 pKa = 3.21GSNDD94 pKa = 2.92VTVGIFSDD102 pKa = 3.94NVEE105 pKa = 4.18LTGFTIKK112 pKa = 10.79NGTNGIMINGSSEE125 pKa = 4.21TIVAEE130 pKa = 3.73NTIIDD135 pKa = 4.16TEE137 pKa = 4.17YY138 pKa = 11.2GIYY141 pKa = 10.29VDD143 pKa = 5.25NKK145 pKa = 9.43SANNTIYY152 pKa = 11.22NNNFLSNIQNAYY164 pKa = 10.38DD165 pKa = 3.57LSTNIWHH172 pKa = 6.27YY173 pKa = 11.43ASNGNYY179 pKa = 8.2WDD181 pKa = 4.85DD182 pKa = 3.68YY183 pKa = 11.01MGIDD187 pKa = 4.92SDD189 pKa = 5.11DD190 pKa = 3.85NGIGDD195 pKa = 3.88TPYY198 pKa = 10.96NISGGNNQDD207 pKa = 3.23IYY209 pKa = 11.17PLMEE213 pKa = 5.96PITEE217 pKa = 4.33KK218 pKa = 10.68PDD220 pKa = 3.37ADD222 pKa = 3.82FTYY225 pKa = 10.62LPSNPTTQDD234 pKa = 3.14VIQFNDD240 pKa = 3.05TSTDD244 pKa = 3.23LDD246 pKa = 4.23GYY248 pKa = 9.41IVSWSWDD255 pKa = 3.48FGDD258 pKa = 5.06GNISMEE264 pKa = 3.98QNATHH269 pKa = 7.41RR270 pKa = 11.84YY271 pKa = 9.32DD272 pKa = 4.3DD273 pKa = 3.71NGVYY277 pKa = 10.57NVTLKK282 pKa = 9.34ITDD285 pKa = 3.42NYY287 pKa = 10.23GATDD291 pKa = 4.42EE292 pKa = 4.63ISRR295 pKa = 11.84QLGVLNVGPTADD307 pKa = 3.73FVYY310 pKa = 10.82SPLNPTDD317 pKa = 3.69LQNVIFTDD325 pKa = 3.59KK326 pKa = 11.27SSDD329 pKa = 3.53LDD331 pKa = 3.65GDD333 pKa = 3.75IVDD336 pKa = 4.68WYY338 pKa = 10.1WDD340 pKa = 3.72LGDD343 pKa = 4.63GNTSKK348 pKa = 10.72LQSPSYY354 pKa = 10.57QYY356 pKa = 11.51GDD358 pKa = 3.15NGTYY362 pKa = 10.06IVTLTITDD370 pKa = 3.85DD371 pKa = 4.35DD372 pKa = 4.53GVSEE376 pKa = 4.41VTSQQISVSNVGPIIDD392 pKa = 3.77FAFFSANVTIIVNDD406 pKa = 3.68PVLFSDD412 pKa = 4.98KK413 pKa = 11.4SEE415 pKa = 4.77DD416 pKa = 3.66LDD418 pKa = 4.28GDD420 pKa = 3.86IVIWSWDD427 pKa = 3.61FGDD430 pKa = 4.97GATSTKK436 pKa = 10.2KK437 pKa = 10.13HH438 pKa = 5.51PTHH441 pKa = 6.65NYY443 pKa = 7.11EE444 pKa = 4.11KK445 pKa = 10.33KK446 pKa = 9.11GTYY449 pKa = 8.8TVTLTVTDD457 pKa = 3.73NDD459 pKa = 4.26GVSSTKK465 pKa = 9.62TKK467 pKa = 10.48YY468 pKa = 8.77ITVSVYY474 pKa = 10.27VEE476 pKa = 3.9PNEE479 pKa = 3.93LVKK482 pKa = 10.95GFSTFDD488 pKa = 2.82IFFVVFLIIMVGLVIFLSRR507 pKa = 11.84KK508 pKa = 9.02YY509 pKa = 10.82GG510 pKa = 3.35
MM1 pKa = 7.34GKK3 pKa = 10.1KK4 pKa = 10.02SVILLLIGLLIFSNSGFNQIEE25 pKa = 3.98NTKK28 pKa = 10.44AVEE31 pKa = 4.05TTFYY35 pKa = 11.31VDD37 pKa = 5.02DD38 pKa = 6.15DD39 pKa = 4.13GLEE42 pKa = 4.37DD43 pKa = 3.8YY44 pKa = 11.85VNIQYY49 pKa = 10.85AIDD52 pKa = 3.66NASIGDD58 pKa = 3.82TVYY61 pKa = 10.86VYY63 pKa = 10.39HH64 pKa = 7.4GYY66 pKa = 10.4YY67 pKa = 9.92KK68 pKa = 10.84EE69 pKa = 3.8NLVINKK75 pKa = 9.52SISLIGEE82 pKa = 4.17DD83 pKa = 4.84KK84 pKa = 11.46GNTTIDD90 pKa = 3.21GSNDD94 pKa = 2.92VTVGIFSDD102 pKa = 3.94NVEE105 pKa = 4.18LTGFTIKK112 pKa = 10.79NGTNGIMINGSSEE125 pKa = 4.21TIVAEE130 pKa = 3.73NTIIDD135 pKa = 4.16TEE137 pKa = 4.17YY138 pKa = 11.2GIYY141 pKa = 10.29VDD143 pKa = 5.25NKK145 pKa = 9.43SANNTIYY152 pKa = 11.22NNNFLSNIQNAYY164 pKa = 10.38DD165 pKa = 3.57LSTNIWHH172 pKa = 6.27YY173 pKa = 11.43ASNGNYY179 pKa = 8.2WDD181 pKa = 4.85DD182 pKa = 3.68YY183 pKa = 11.01MGIDD187 pKa = 4.92SDD189 pKa = 5.11DD190 pKa = 3.85NGIGDD195 pKa = 3.88TPYY198 pKa = 10.96NISGGNNQDD207 pKa = 3.23IYY209 pKa = 11.17PLMEE213 pKa = 5.96PITEE217 pKa = 4.33KK218 pKa = 10.68PDD220 pKa = 3.37ADD222 pKa = 3.82FTYY225 pKa = 10.62LPSNPTTQDD234 pKa = 3.14VIQFNDD240 pKa = 3.05TSTDD244 pKa = 3.23LDD246 pKa = 4.23GYY248 pKa = 9.41IVSWSWDD255 pKa = 3.48FGDD258 pKa = 5.06GNISMEE264 pKa = 3.98QNATHH269 pKa = 7.41RR270 pKa = 11.84YY271 pKa = 9.32DD272 pKa = 4.3DD273 pKa = 3.71NGVYY277 pKa = 10.57NVTLKK282 pKa = 9.34ITDD285 pKa = 3.42NYY287 pKa = 10.23GATDD291 pKa = 4.42EE292 pKa = 4.63ISRR295 pKa = 11.84QLGVLNVGPTADD307 pKa = 3.73FVYY310 pKa = 10.82SPLNPTDD317 pKa = 3.69LQNVIFTDD325 pKa = 3.59KK326 pKa = 11.27SSDD329 pKa = 3.53LDD331 pKa = 3.65GDD333 pKa = 3.75IVDD336 pKa = 4.68WYY338 pKa = 10.1WDD340 pKa = 3.72LGDD343 pKa = 4.63GNTSKK348 pKa = 10.72LQSPSYY354 pKa = 10.57QYY356 pKa = 11.51GDD358 pKa = 3.15NGTYY362 pKa = 10.06IVTLTITDD370 pKa = 3.85DD371 pKa = 4.35DD372 pKa = 4.53GVSEE376 pKa = 4.41VTSQQISVSNVGPIIDD392 pKa = 3.77FAFFSANVTIIVNDD406 pKa = 3.68PVLFSDD412 pKa = 4.98KK413 pKa = 11.4SEE415 pKa = 4.77DD416 pKa = 3.66LDD418 pKa = 4.28GDD420 pKa = 3.86IVIWSWDD427 pKa = 3.61FGDD430 pKa = 4.97GATSTKK436 pKa = 10.2KK437 pKa = 10.13HH438 pKa = 5.51PTHH441 pKa = 6.65NYY443 pKa = 7.11EE444 pKa = 4.11KK445 pKa = 10.33KK446 pKa = 9.11GTYY449 pKa = 8.8TVTLTVTDD457 pKa = 3.73NDD459 pKa = 4.26GVSSTKK465 pKa = 9.62TKK467 pKa = 10.48YY468 pKa = 8.77ITVSVYY474 pKa = 10.27VEE476 pKa = 3.9PNEE479 pKa = 3.93LVKK482 pKa = 10.95GFSTFDD488 pKa = 2.82IFFVVFLIIMVGLVIFLSRR507 pKa = 11.84KK508 pKa = 9.02YY509 pKa = 10.82GG510 pKa = 3.35
Molecular weight: 56.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N6VWM2|N6VWM2_9ARCH Acetyltransferase OS=Thermoplasmatales archaeon SCGC AB-539-C06 OX=1242690 GN=MBGDC06_00010 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.23IKK4 pKa = 9.33TRR6 pKa = 11.84KK7 pKa = 8.94KK8 pKa = 8.48VYY10 pKa = 10.4GRR12 pKa = 11.84IKK14 pKa = 9.73GCEE17 pKa = 3.3RR18 pKa = 11.84CGRR21 pKa = 11.84KK22 pKa = 9.31RR23 pKa = 11.84GLIRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.8GLHH32 pKa = 7.31LCRR35 pKa = 11.84QCFRR39 pKa = 11.84EE40 pKa = 3.94KK41 pKa = 10.93AEE43 pKa = 4.21EE44 pKa = 3.99IGFKK48 pKa = 10.37KK49 pKa = 10.85YY50 pKa = 10.21SS51 pKa = 3.24
MM1 pKa = 7.74KK2 pKa = 10.23IKK4 pKa = 9.33TRR6 pKa = 11.84KK7 pKa = 8.94KK8 pKa = 8.48VYY10 pKa = 10.4GRR12 pKa = 11.84IKK14 pKa = 9.73GCEE17 pKa = 3.3RR18 pKa = 11.84CGRR21 pKa = 11.84KK22 pKa = 9.31RR23 pKa = 11.84GLIRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.8GLHH32 pKa = 7.31LCRR35 pKa = 11.84QCFRR39 pKa = 11.84EE40 pKa = 3.94KK41 pKa = 10.93AEE43 pKa = 4.21EE44 pKa = 3.99IGFKK48 pKa = 10.37KK49 pKa = 10.85YY50 pKa = 10.21SS51 pKa = 3.24
Molecular weight: 6.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
164750 |
22 |
1147 |
211.2 |
23.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.658 ± 0.125 | 1.339 ± 0.05 |
5.845 ± 0.1 | 6.778 ± 0.116 |
4.522 ± 0.081 | 6.987 ± 0.11 |
1.818 ± 0.038 | 9.249 ± 0.12 |
8.407 ± 0.147 | 8.456 ± 0.117 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.562 ± 0.049 | 5.17 ± 0.106 |
3.635 ± 0.058 | 2.452 ± 0.05 |
3.775 ± 0.074 | 6.281 ± 0.078 |
5.423 ± 0.095 | 6.608 ± 0.089 |
1.212 ± 0.042 | 3.824 ± 0.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
