
Chimpanzee herpesvirus strain 105640
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus; Panine alphaherpesvirus 3
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
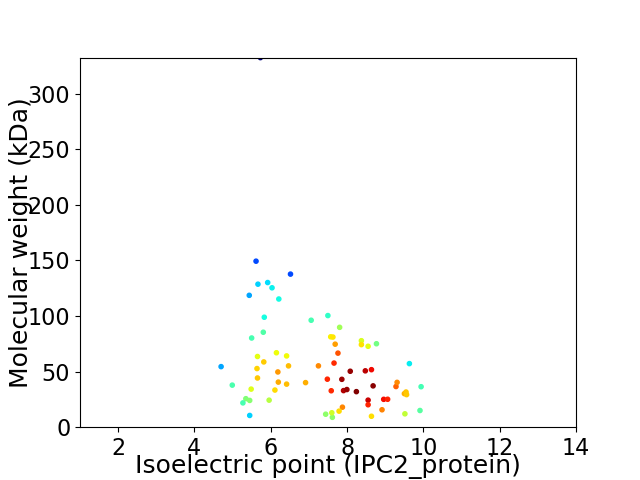
Virtual 2D-PAGE plot for 74 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9MG54|K9MG54_9ALPH DNA primase OS=Chimpanzee herpesvirus strain 105640 OX=332937 GN=UL52 PE=3 SV=1
MM1 pKa = 7.97DD2 pKa = 6.11LLVDD6 pKa = 5.22DD7 pKa = 5.74LFADD11 pKa = 4.0ADD13 pKa = 4.47GVSSPPTRR21 pKa = 11.84PAGGPKK27 pKa = 8.46NTPAAPPLYY36 pKa = 9.66ATGRR40 pKa = 11.84LSQAQLMPSPPMPVPPAALFNRR62 pKa = 11.84LLDD65 pKa = 4.24DD66 pKa = 4.82LGFSAGPALCTMLDD80 pKa = 3.31TWNEE84 pKa = 3.9DD85 pKa = 3.45LFSGFPTNADD95 pKa = 3.44MYY97 pKa = 10.28RR98 pKa = 11.84EE99 pKa = 4.25CKK101 pKa = 9.87FLSTLPSDD109 pKa = 3.83VIEE112 pKa = 4.53WGDD115 pKa = 3.35AYY117 pKa = 11.22VPEE120 pKa = 5.04RR121 pKa = 11.84SQIDD125 pKa = 3.2IRR127 pKa = 11.84AHH129 pKa = 5.95GDD131 pKa = 3.29VAFPTLPATRR141 pKa = 11.84DD142 pKa = 3.5GLASYY147 pKa = 9.89YY148 pKa = 10.68EE149 pKa = 4.01AMAQFFRR156 pKa = 11.84GEE158 pKa = 3.49LRR160 pKa = 11.84ARR162 pKa = 11.84EE163 pKa = 3.85EE164 pKa = 4.07SYY166 pKa = 9.67RR167 pKa = 11.84TVLANFCSALYY178 pKa = 9.82RR179 pKa = 11.84YY180 pKa = 9.56LRR182 pKa = 11.84ASVRR186 pKa = 11.84QLHH189 pKa = 5.87RR190 pKa = 11.84QAHH193 pKa = 4.64MRR195 pKa = 11.84GRR197 pKa = 11.84DD198 pKa = 3.1RR199 pKa = 11.84DD200 pKa = 3.78LRR202 pKa = 11.84EE203 pKa = 3.82MLRR206 pKa = 11.84TTIADD211 pKa = 3.77RR212 pKa = 11.84YY213 pKa = 8.15YY214 pKa = 11.05RR215 pKa = 11.84EE216 pKa = 3.99TARR219 pKa = 11.84LARR222 pKa = 11.84VLFLHH227 pKa = 7.13LYY229 pKa = 10.43LFLTRR234 pKa = 11.84EE235 pKa = 4.38ILWAAYY241 pKa = 9.3AEE243 pKa = 4.24QMMRR247 pKa = 11.84PDD249 pKa = 5.09LFDD252 pKa = 4.58GLCCDD257 pKa = 4.14LEE259 pKa = 4.27SWRR262 pKa = 11.84QLACLFQPLMFINGSLTVRR281 pKa = 11.84GVPVEE286 pKa = 3.73ARR288 pKa = 11.84RR289 pKa = 11.84LRR291 pKa = 11.84EE292 pKa = 3.74LNHH295 pKa = 6.26IRR297 pKa = 11.84EE298 pKa = 4.46HH299 pKa = 6.68LNLPLIRR306 pKa = 11.84SAAAEE311 pKa = 4.22EE312 pKa = 4.44PGAPLTTPPVLQGNQARR329 pKa = 11.84ASGYY333 pKa = 10.54FMLLIRR339 pKa = 11.84AKK341 pKa = 10.0LDD343 pKa = 3.46SYY345 pKa = 11.74SSVATSEE352 pKa = 4.58GEE354 pKa = 4.25TVMRR358 pKa = 11.84EE359 pKa = 3.68HH360 pKa = 7.11AYY362 pKa = 10.56SRR364 pKa = 11.84GRR366 pKa = 11.84HH367 pKa = 4.78RR368 pKa = 11.84NNYY371 pKa = 8.94GSTIEE376 pKa = 4.57GLLDD380 pKa = 5.1LPDD383 pKa = 5.66DD384 pKa = 5.23DD385 pKa = 5.87DD386 pKa = 5.94APAEE390 pKa = 4.39AGLVAPRR397 pKa = 11.84MSFLSAGQHH406 pKa = 5.22PRR408 pKa = 11.84RR409 pKa = 11.84LSTAPLTDD417 pKa = 3.1VSLGDD422 pKa = 3.68EE423 pKa = 4.16LRR425 pKa = 11.84LGGEE429 pKa = 4.45EE430 pKa = 4.1VDD432 pKa = 3.85MTPVDD437 pKa = 3.71ALDD440 pKa = 5.63DD441 pKa = 4.22FDD443 pKa = 6.99LDD445 pKa = 3.58MLGDD449 pKa = 3.84VEE451 pKa = 5.02FPSPGMTHH459 pKa = 7.23DD460 pKa = 3.94PVPYY464 pKa = 10.11GALDD468 pKa = 3.42VADD471 pKa = 5.25FEE473 pKa = 4.53FEE475 pKa = 4.78QMFTDD480 pKa = 4.69ALGIDD485 pKa = 4.2DD486 pKa = 4.99FGGG489 pKa = 3.36
MM1 pKa = 7.97DD2 pKa = 6.11LLVDD6 pKa = 5.22DD7 pKa = 5.74LFADD11 pKa = 4.0ADD13 pKa = 4.47GVSSPPTRR21 pKa = 11.84PAGGPKK27 pKa = 8.46NTPAAPPLYY36 pKa = 9.66ATGRR40 pKa = 11.84LSQAQLMPSPPMPVPPAALFNRR62 pKa = 11.84LLDD65 pKa = 4.24DD66 pKa = 4.82LGFSAGPALCTMLDD80 pKa = 3.31TWNEE84 pKa = 3.9DD85 pKa = 3.45LFSGFPTNADD95 pKa = 3.44MYY97 pKa = 10.28RR98 pKa = 11.84EE99 pKa = 4.25CKK101 pKa = 9.87FLSTLPSDD109 pKa = 3.83VIEE112 pKa = 4.53WGDD115 pKa = 3.35AYY117 pKa = 11.22VPEE120 pKa = 5.04RR121 pKa = 11.84SQIDD125 pKa = 3.2IRR127 pKa = 11.84AHH129 pKa = 5.95GDD131 pKa = 3.29VAFPTLPATRR141 pKa = 11.84DD142 pKa = 3.5GLASYY147 pKa = 9.89YY148 pKa = 10.68EE149 pKa = 4.01AMAQFFRR156 pKa = 11.84GEE158 pKa = 3.49LRR160 pKa = 11.84ARR162 pKa = 11.84EE163 pKa = 3.85EE164 pKa = 4.07SYY166 pKa = 9.67RR167 pKa = 11.84TVLANFCSALYY178 pKa = 9.82RR179 pKa = 11.84YY180 pKa = 9.56LRR182 pKa = 11.84ASVRR186 pKa = 11.84QLHH189 pKa = 5.87RR190 pKa = 11.84QAHH193 pKa = 4.64MRR195 pKa = 11.84GRR197 pKa = 11.84DD198 pKa = 3.1RR199 pKa = 11.84DD200 pKa = 3.78LRR202 pKa = 11.84EE203 pKa = 3.82MLRR206 pKa = 11.84TTIADD211 pKa = 3.77RR212 pKa = 11.84YY213 pKa = 8.15YY214 pKa = 11.05RR215 pKa = 11.84EE216 pKa = 3.99TARR219 pKa = 11.84LARR222 pKa = 11.84VLFLHH227 pKa = 7.13LYY229 pKa = 10.43LFLTRR234 pKa = 11.84EE235 pKa = 4.38ILWAAYY241 pKa = 9.3AEE243 pKa = 4.24QMMRR247 pKa = 11.84PDD249 pKa = 5.09LFDD252 pKa = 4.58GLCCDD257 pKa = 4.14LEE259 pKa = 4.27SWRR262 pKa = 11.84QLACLFQPLMFINGSLTVRR281 pKa = 11.84GVPVEE286 pKa = 3.73ARR288 pKa = 11.84RR289 pKa = 11.84LRR291 pKa = 11.84EE292 pKa = 3.74LNHH295 pKa = 6.26IRR297 pKa = 11.84EE298 pKa = 4.46HH299 pKa = 6.68LNLPLIRR306 pKa = 11.84SAAAEE311 pKa = 4.22EE312 pKa = 4.44PGAPLTTPPVLQGNQARR329 pKa = 11.84ASGYY333 pKa = 10.54FMLLIRR339 pKa = 11.84AKK341 pKa = 10.0LDD343 pKa = 3.46SYY345 pKa = 11.74SSVATSEE352 pKa = 4.58GEE354 pKa = 4.25TVMRR358 pKa = 11.84EE359 pKa = 3.68HH360 pKa = 7.11AYY362 pKa = 10.56SRR364 pKa = 11.84GRR366 pKa = 11.84HH367 pKa = 4.78RR368 pKa = 11.84NNYY371 pKa = 8.94GSTIEE376 pKa = 4.57GLLDD380 pKa = 5.1LPDD383 pKa = 5.66DD384 pKa = 5.23DD385 pKa = 5.87DD386 pKa = 5.94APAEE390 pKa = 4.39AGLVAPRR397 pKa = 11.84MSFLSAGQHH406 pKa = 5.22PRR408 pKa = 11.84RR409 pKa = 11.84LSTAPLTDD417 pKa = 3.1VSLGDD422 pKa = 3.68EE423 pKa = 4.16LRR425 pKa = 11.84LGGEE429 pKa = 4.45EE430 pKa = 4.1VDD432 pKa = 3.85MTPVDD437 pKa = 3.71ALDD440 pKa = 5.63DD441 pKa = 4.22FDD443 pKa = 6.99LDD445 pKa = 3.58MLGDD449 pKa = 3.84VEE451 pKa = 5.02FPSPGMTHH459 pKa = 7.23DD460 pKa = 3.94PVPYY464 pKa = 10.11GALDD468 pKa = 3.42VADD471 pKa = 5.25FEE473 pKa = 4.53FEE475 pKa = 4.78QMFTDD480 pKa = 4.69ALGIDD485 pKa = 4.2DD486 pKa = 4.99FGGG489 pKa = 3.36
Molecular weight: 54.49 kDa
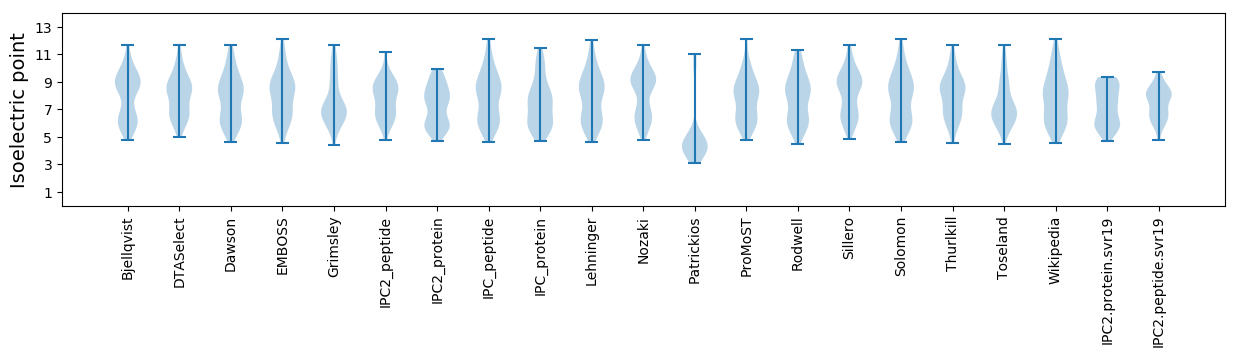
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q3C1Z5|Q3C1Z5_9ALPH Thymidine kinase OS=Chimpanzee herpesvirus strain 105640 OX=332937 GN=UL23 PE=3 SV=1
MM1 pKa = 7.67AARR4 pKa = 11.84TSSLSEE10 pKa = 3.72RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84VLLAGVRR20 pKa = 11.84SHH22 pKa = 5.27TRR24 pKa = 11.84FYY26 pKa = 11.02KK27 pKa = 10.32AFARR31 pKa = 11.84EE32 pKa = 3.85VRR34 pKa = 11.84EE35 pKa = 4.03FNATRR40 pKa = 11.84ICGTLLTLMSGSLQGRR56 pKa = 11.84SLFEE60 pKa = 3.81ATRR63 pKa = 11.84VTLICEE69 pKa = 3.9VDD71 pKa = 3.8LGPRR75 pKa = 11.84RR76 pKa = 11.84PDD78 pKa = 3.88CICVFEE84 pKa = 4.16FANDD88 pKa = 3.31KK89 pKa = 9.57TLGGVCVILEE99 pKa = 4.58LKK101 pKa = 7.05TCKK104 pKa = 10.39YY105 pKa = 10.07ISSGDD110 pKa = 3.52TASKK114 pKa = 10.46RR115 pKa = 11.84EE116 pKa = 3.79QRR118 pKa = 11.84TTGMKK123 pKa = 9.7QLRR126 pKa = 11.84HH127 pKa = 4.98SVKK130 pKa = 10.24LLQSLAPPGDD140 pKa = 3.39KK141 pKa = 10.35VVYY144 pKa = 9.85LCPILVFVAQRR155 pKa = 11.84TLRR158 pKa = 11.84VSRR161 pKa = 11.84VTRR164 pKa = 11.84LVPQKK169 pKa = 10.43ISGNITAAVRR179 pKa = 11.84MLQSLSTYY187 pKa = 9.97AVPPEE192 pKa = 3.78PRR194 pKa = 11.84TRR196 pKa = 11.84QTRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84GAAGGRR207 pKa = 11.84PQRR210 pKa = 11.84PPSPARR216 pKa = 11.84APDD219 pKa = 3.52GTEE222 pKa = 3.93GHH224 pKa = 7.19PSPPEE229 pKa = 3.78SDD231 pKa = 3.3TPSAVVVGVAAEE243 pKa = 4.32GGGVLQKK250 pKa = 10.28IAALFCVPVAAKK262 pKa = 10.14SKK264 pKa = 10.17PRR266 pKa = 11.84TKK268 pKa = 10.42TEE270 pKa = 3.55
MM1 pKa = 7.67AARR4 pKa = 11.84TSSLSEE10 pKa = 3.72RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84VLLAGVRR20 pKa = 11.84SHH22 pKa = 5.27TRR24 pKa = 11.84FYY26 pKa = 11.02KK27 pKa = 10.32AFARR31 pKa = 11.84EE32 pKa = 3.85VRR34 pKa = 11.84EE35 pKa = 4.03FNATRR40 pKa = 11.84ICGTLLTLMSGSLQGRR56 pKa = 11.84SLFEE60 pKa = 3.81ATRR63 pKa = 11.84VTLICEE69 pKa = 3.9VDD71 pKa = 3.8LGPRR75 pKa = 11.84RR76 pKa = 11.84PDD78 pKa = 3.88CICVFEE84 pKa = 4.16FANDD88 pKa = 3.31KK89 pKa = 9.57TLGGVCVILEE99 pKa = 4.58LKK101 pKa = 7.05TCKK104 pKa = 10.39YY105 pKa = 10.07ISSGDD110 pKa = 3.52TASKK114 pKa = 10.46RR115 pKa = 11.84EE116 pKa = 3.79QRR118 pKa = 11.84TTGMKK123 pKa = 9.7QLRR126 pKa = 11.84HH127 pKa = 4.98SVKK130 pKa = 10.24LLQSLAPPGDD140 pKa = 3.39KK141 pKa = 10.35VVYY144 pKa = 9.85LCPILVFVAQRR155 pKa = 11.84TLRR158 pKa = 11.84VSRR161 pKa = 11.84VTRR164 pKa = 11.84LVPQKK169 pKa = 10.43ISGNITAAVRR179 pKa = 11.84MLQSLSTYY187 pKa = 9.97AVPPEE192 pKa = 3.78PRR194 pKa = 11.84TRR196 pKa = 11.84QTRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84GAAGGRR207 pKa = 11.84PQRR210 pKa = 11.84PPSPARR216 pKa = 11.84APDD219 pKa = 3.52GTEE222 pKa = 3.93GHH224 pKa = 7.19PSPPEE229 pKa = 3.78SDD231 pKa = 3.3TPSAVVVGVAAEE243 pKa = 4.32GGGVLQKK250 pKa = 10.28IAALFCVPVAAKK262 pKa = 10.14SKK264 pKa = 10.17PRR266 pKa = 11.84TKK268 pKa = 10.42TEE270 pKa = 3.55
Molecular weight: 29.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
39168 |
87 |
3129 |
529.3 |
57.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.906 ± 0.422 | 1.841 ± 0.112 |
5.162 ± 0.13 | 4.836 ± 0.139 |
3.424 ± 0.171 | 7.721 ± 0.225 |
2.66 ± 0.103 | 2.87 ± 0.153 |
1.665 ± 0.141 | 9.643 ± 0.224 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.67 ± 0.094 | 2.211 ± 0.149 |
8.826 ± 0.471 | 2.969 ± 0.133 |
8.609 ± 0.181 | 6.23 ± 0.206 |
5.9 ± 0.155 | 7.177 ± 0.192 |
1.151 ± 0.069 | 2.53 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
