
Nonomuraea wenchangensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Streptosporangiaceae; Nonomuraea
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
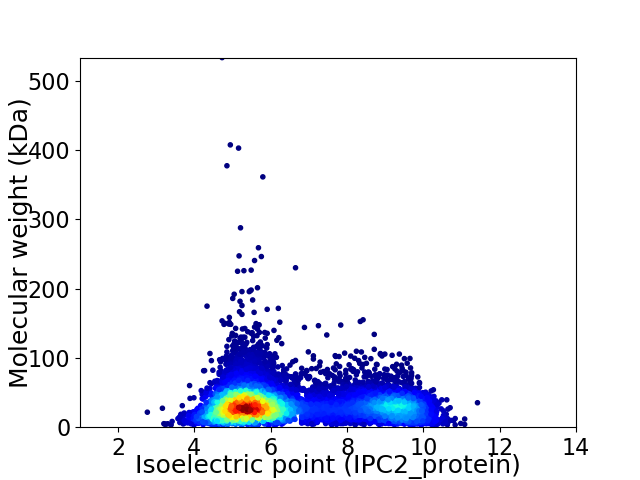
Virtual 2D-PAGE plot for 9568 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
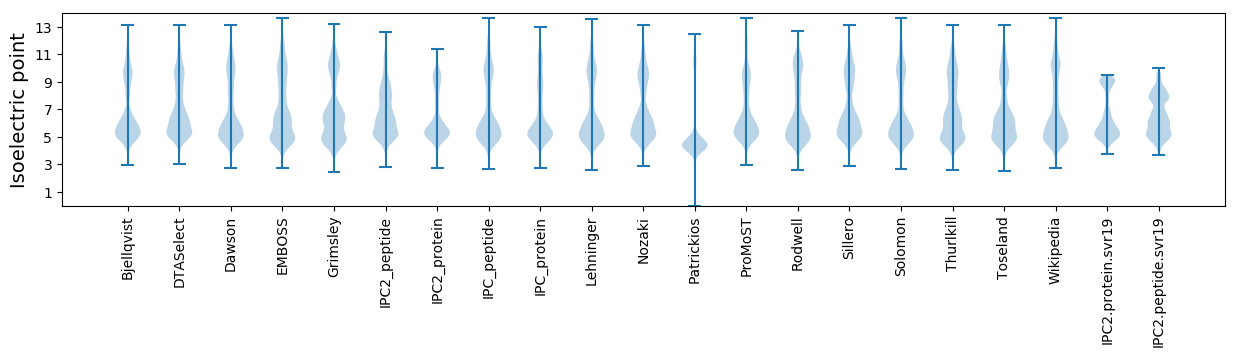
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0LPK3|A0A1I0LPK3_9ACTN Fatty acid CoA ligase FadD32 OS=Nonomuraea wenchangensis OX=568860 GN=SAMN05421811_121128 PE=4 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84VRR4 pKa = 11.84VDD6 pKa = 3.31LTLCQTHH13 pKa = 6.04AQCVFAAPEE22 pKa = 4.1VFALDD27 pKa = 4.56DD28 pKa = 4.75DD29 pKa = 5.23DD30 pKa = 5.0EE31 pKa = 4.63LVYY34 pKa = 10.95DD35 pKa = 4.7ATPGDD40 pKa = 4.07ASWPAVEE47 pKa = 4.36QAARR51 pKa = 11.84ACPVQAIFLDD61 pKa = 4.36GDD63 pKa = 3.63
MM1 pKa = 7.88RR2 pKa = 11.84VRR4 pKa = 11.84VDD6 pKa = 3.31LTLCQTHH13 pKa = 6.04AQCVFAAPEE22 pKa = 4.1VFALDD27 pKa = 4.56DD28 pKa = 4.75DD29 pKa = 5.23DD30 pKa = 5.0EE31 pKa = 4.63LVYY34 pKa = 10.95DD35 pKa = 4.7ATPGDD40 pKa = 4.07ASWPAVEE47 pKa = 4.36QAARR51 pKa = 11.84ACPVQAIFLDD61 pKa = 4.36GDD63 pKa = 3.63
Molecular weight: 6.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0LMZ2|A0A1I0LMZ2_9ACTN RNA polymerase sigma-70 factor ECF subfamily OS=Nonomuraea wenchangensis OX=568860 GN=SAMN05421811_12096 PE=3 SV=1
MM1 pKa = 6.33QWRR4 pKa = 11.84RR5 pKa = 11.84SGYY8 pKa = 8.05TSSLRR13 pKa = 11.84SSASRR18 pKa = 11.84ARR20 pKa = 11.84SSWPSSKK27 pKa = 9.76RR28 pKa = 11.84WGSSSVRR35 pKa = 11.84RR36 pKa = 11.84PPPSRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84WSAGSPKK50 pKa = 9.92LWAHH54 pKa = 6.11RR55 pKa = 11.84RR56 pKa = 11.84VGPPGAADD64 pKa = 3.37SRR66 pKa = 11.84PTGRR70 pKa = 11.84SRR72 pKa = 11.84QGLRR76 pKa = 11.84TGRR79 pKa = 11.84RR80 pKa = 11.84KK81 pKa = 9.78ARR83 pKa = 11.84PATVPGRR90 pKa = 11.84PRR92 pKa = 11.84CLLLRR97 pKa = 11.84VRR99 pKa = 11.84APGPVRR105 pKa = 11.84VRR107 pKa = 11.84PRR109 pKa = 11.84RR110 pKa = 11.84AALRR114 pKa = 11.84VRR116 pKa = 11.84RR117 pKa = 11.84GPRR120 pKa = 11.84ARR122 pKa = 11.84RR123 pKa = 11.84VRR125 pKa = 11.84KK126 pKa = 9.22VLRR129 pKa = 11.84AAASPARR136 pKa = 11.84RR137 pKa = 11.84FPCLTSSSRR146 pKa = 11.84RR147 pKa = 11.84PRR149 pKa = 11.84AVRR152 pKa = 11.84RR153 pKa = 11.84PRR155 pKa = 11.84LRR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84ASRR162 pKa = 11.84AARR165 pKa = 11.84RR166 pKa = 11.84PGRR169 pKa = 11.84PRR171 pKa = 11.84VRR173 pKa = 11.84VPARR177 pKa = 11.84LRR179 pKa = 11.84VRR181 pKa = 11.84VPAPSRR187 pKa = 11.84ARR189 pKa = 11.84VPPSRR194 pKa = 11.84AAAARR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84VLAARR208 pKa = 11.84PPAAARR214 pKa = 11.84VPAPSRR220 pKa = 11.84ARR222 pKa = 11.84VARR225 pKa = 11.84VRR227 pKa = 11.84ATTRR231 pKa = 11.84SRR233 pKa = 11.84PTPAAWARR241 pKa = 11.84RR242 pKa = 11.84PGRR245 pKa = 11.84SVRR248 pKa = 11.84AAASVTVAPARR259 pKa = 11.84VASAVTARR267 pKa = 11.84RR268 pKa = 11.84ATARR272 pKa = 11.84CRR274 pKa = 11.84VRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84VAAATGPLPARR289 pKa = 11.84VPVRR293 pKa = 11.84PPAARR298 pKa = 11.84VRR300 pKa = 11.84VPAPVAPVVRR310 pKa = 11.84VPVVRR315 pKa = 11.84VPTRR319 pKa = 3.06
MM1 pKa = 6.33QWRR4 pKa = 11.84RR5 pKa = 11.84SGYY8 pKa = 8.05TSSLRR13 pKa = 11.84SSASRR18 pKa = 11.84ARR20 pKa = 11.84SSWPSSKK27 pKa = 9.76RR28 pKa = 11.84WGSSSVRR35 pKa = 11.84RR36 pKa = 11.84PPPSRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84WSAGSPKK50 pKa = 9.92LWAHH54 pKa = 6.11RR55 pKa = 11.84RR56 pKa = 11.84VGPPGAADD64 pKa = 3.37SRR66 pKa = 11.84PTGRR70 pKa = 11.84SRR72 pKa = 11.84QGLRR76 pKa = 11.84TGRR79 pKa = 11.84RR80 pKa = 11.84KK81 pKa = 9.78ARR83 pKa = 11.84PATVPGRR90 pKa = 11.84PRR92 pKa = 11.84CLLLRR97 pKa = 11.84VRR99 pKa = 11.84APGPVRR105 pKa = 11.84VRR107 pKa = 11.84PRR109 pKa = 11.84RR110 pKa = 11.84AALRR114 pKa = 11.84VRR116 pKa = 11.84RR117 pKa = 11.84GPRR120 pKa = 11.84ARR122 pKa = 11.84RR123 pKa = 11.84VRR125 pKa = 11.84KK126 pKa = 9.22VLRR129 pKa = 11.84AAASPARR136 pKa = 11.84RR137 pKa = 11.84FPCLTSSSRR146 pKa = 11.84RR147 pKa = 11.84PRR149 pKa = 11.84AVRR152 pKa = 11.84RR153 pKa = 11.84PRR155 pKa = 11.84LRR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84ASRR162 pKa = 11.84AARR165 pKa = 11.84RR166 pKa = 11.84PGRR169 pKa = 11.84PRR171 pKa = 11.84VRR173 pKa = 11.84VPARR177 pKa = 11.84LRR179 pKa = 11.84VRR181 pKa = 11.84VPAPSRR187 pKa = 11.84ARR189 pKa = 11.84VPPSRR194 pKa = 11.84AAAARR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84VLAARR208 pKa = 11.84PPAAARR214 pKa = 11.84VPAPSRR220 pKa = 11.84ARR222 pKa = 11.84VARR225 pKa = 11.84VRR227 pKa = 11.84ATTRR231 pKa = 11.84SRR233 pKa = 11.84PTPAAWARR241 pKa = 11.84RR242 pKa = 11.84PGRR245 pKa = 11.84SVRR248 pKa = 11.84AAASVTVAPARR259 pKa = 11.84VASAVTARR267 pKa = 11.84RR268 pKa = 11.84ATARR272 pKa = 11.84CRR274 pKa = 11.84VRR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84VAAATGPLPARR289 pKa = 11.84VPVRR293 pKa = 11.84PPAARR298 pKa = 11.84VRR300 pKa = 11.84VPAPVAPVVRR310 pKa = 11.84VPVVRR315 pKa = 11.84VPTRR319 pKa = 3.06
Molecular weight: 35.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3041378 |
27 |
5134 |
317.9 |
34.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.689 ± 0.041 | 0.783 ± 0.007 |
5.71 ± 0.019 | 5.737 ± 0.025 |
2.771 ± 0.014 | 9.454 ± 0.027 |
2.205 ± 0.012 | 3.317 ± 0.018 |
1.958 ± 0.019 | 10.778 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.853 ± 0.01 | 1.725 ± 0.014 |
6.13 ± 0.021 | 2.618 ± 0.017 |
8.337 ± 0.028 | 4.782 ± 0.017 |
5.716 ± 0.02 | 8.717 ± 0.026 |
1.595 ± 0.012 | 2.126 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
