
Sanxia Water Strider Virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Nairoviridae; Striwavirus; Strider striwavirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
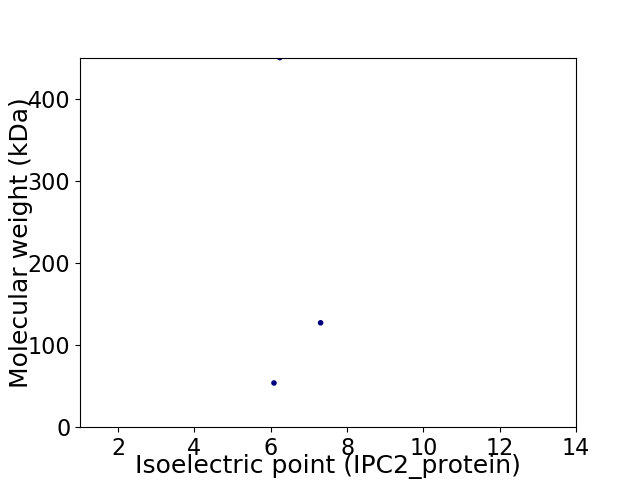
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
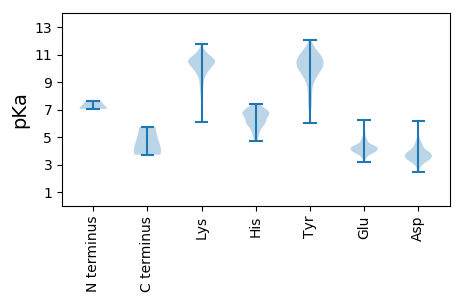
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KS14|A0A0B5KS14_9VIRU Glycoprotein G2 OS=Sanxia Water Strider Virus 1 OX=1608060 GN=G PE=4 SV=1
MM1 pKa = 7.63AAPAPFNVITSYY13 pKa = 9.26EE14 pKa = 4.18TASVSVNALRR24 pKa = 11.84TVSVNITSCPTFLNAFPDD42 pKa = 3.37NYY44 pKa = 10.96NNYY47 pKa = 7.96IQKK50 pKa = 9.93INAMRR55 pKa = 11.84AASPKK60 pKa = 9.72EE61 pKa = 3.93KK62 pKa = 10.41NAVVADD68 pKa = 4.01MVTEE72 pKa = 4.37TLRR75 pKa = 11.84DD76 pKa = 3.44ACPAVAQFYY85 pKa = 10.74CLGQGFVTRR94 pKa = 11.84SRR96 pKa = 11.84AFFSTDD102 pKa = 2.75PSVQVLNYY110 pKa = 9.77DD111 pKa = 3.72AYY113 pKa = 9.94IATGTQVTDD122 pKa = 3.64PKK124 pKa = 11.08EE125 pKa = 3.9IDD127 pKa = 4.35DD128 pKa = 4.37YY129 pKa = 10.86NHH131 pKa = 6.81CVDD134 pKa = 3.84SYY136 pKa = 9.91RR137 pKa = 11.84TYY139 pKa = 10.21MVNGGFALNNEE150 pKa = 4.32LATLRR155 pKa = 11.84GAVKK159 pKa = 9.28NTINASGNLVPLQVMLRR176 pKa = 11.84EE177 pKa = 4.34MIEE180 pKa = 3.76KK181 pKa = 10.32RR182 pKa = 11.84RR183 pKa = 11.84FNLGGTSAGAEE194 pKa = 4.05HH195 pKa = 6.35LRR197 pKa = 11.84EE198 pKa = 4.0VRR200 pKa = 11.84EE201 pKa = 4.11ALSGDD206 pKa = 3.96LDD208 pKa = 3.73QKK210 pKa = 11.01IIFPFSKK217 pKa = 10.23SEE219 pKa = 3.83RR220 pKa = 11.84KK221 pKa = 9.4AVGFSFSNQINNRR234 pKa = 11.84AEE236 pKa = 3.92RR237 pKa = 11.84NEE239 pKa = 3.86GGRR242 pKa = 11.84CLLASTAVAKK252 pKa = 8.2DD253 pKa = 3.54TQLEE257 pKa = 4.09GDD259 pKa = 4.41LLNTITRR266 pKa = 11.84KK267 pKa = 8.92FHH269 pKa = 7.38AMPQDD274 pKa = 3.31HH275 pKa = 7.01VNYY278 pKa = 10.24QLFDD282 pKa = 4.94RR283 pKa = 11.84IIRR286 pKa = 11.84EE287 pKa = 4.09ANAILDD293 pKa = 4.16PEE295 pKa = 4.76DD296 pKa = 3.47VDD298 pKa = 4.14AADD301 pKa = 3.93GAGFLSQRR309 pKa = 11.84SNIDD313 pKa = 3.55PLWLTFYY320 pKa = 10.8FAFKK324 pKa = 10.42VIKK327 pKa = 10.54LSVADD332 pKa = 4.51FYY334 pKa = 11.39KK335 pKa = 9.63WQRR338 pKa = 11.84AIYY341 pKa = 7.33ATCRR345 pKa = 11.84KK346 pKa = 9.36IRR348 pKa = 11.84GKK350 pKa = 10.54EE351 pKa = 3.74NIRR354 pKa = 11.84AVLATMLDD362 pKa = 3.67LPGVQDD368 pKa = 3.7TLGKK372 pKa = 9.47MSQSTEE378 pKa = 3.18IHH380 pKa = 5.25TPPWVMHH387 pKa = 6.13FSRR390 pKa = 11.84LYY392 pKa = 10.67DD393 pKa = 3.3IAAVICPCDD402 pKa = 3.78AYY404 pKa = 11.33NVAATIQTPAISWRR418 pKa = 11.84YY419 pKa = 9.69LFTVNPAVKK428 pKa = 10.38GFISYY433 pKa = 7.81LTTLNAVEE441 pKa = 4.27KK442 pKa = 9.45TADD445 pKa = 3.58EE446 pKa = 4.2TSEE449 pKa = 3.66IHH451 pKa = 5.75QSEE454 pKa = 4.6RR455 pKa = 11.84MLQQFILGRR464 pKa = 11.84NSPLANTNHH473 pKa = 6.04LTGNALDD480 pKa = 3.82VTIVGG485 pKa = 4.02
MM1 pKa = 7.63AAPAPFNVITSYY13 pKa = 9.26EE14 pKa = 4.18TASVSVNALRR24 pKa = 11.84TVSVNITSCPTFLNAFPDD42 pKa = 3.37NYY44 pKa = 10.96NNYY47 pKa = 7.96IQKK50 pKa = 9.93INAMRR55 pKa = 11.84AASPKK60 pKa = 9.72EE61 pKa = 3.93KK62 pKa = 10.41NAVVADD68 pKa = 4.01MVTEE72 pKa = 4.37TLRR75 pKa = 11.84DD76 pKa = 3.44ACPAVAQFYY85 pKa = 10.74CLGQGFVTRR94 pKa = 11.84SRR96 pKa = 11.84AFFSTDD102 pKa = 2.75PSVQVLNYY110 pKa = 9.77DD111 pKa = 3.72AYY113 pKa = 9.94IATGTQVTDD122 pKa = 3.64PKK124 pKa = 11.08EE125 pKa = 3.9IDD127 pKa = 4.35DD128 pKa = 4.37YY129 pKa = 10.86NHH131 pKa = 6.81CVDD134 pKa = 3.84SYY136 pKa = 9.91RR137 pKa = 11.84TYY139 pKa = 10.21MVNGGFALNNEE150 pKa = 4.32LATLRR155 pKa = 11.84GAVKK159 pKa = 9.28NTINASGNLVPLQVMLRR176 pKa = 11.84EE177 pKa = 4.34MIEE180 pKa = 3.76KK181 pKa = 10.32RR182 pKa = 11.84RR183 pKa = 11.84FNLGGTSAGAEE194 pKa = 4.05HH195 pKa = 6.35LRR197 pKa = 11.84EE198 pKa = 4.0VRR200 pKa = 11.84EE201 pKa = 4.11ALSGDD206 pKa = 3.96LDD208 pKa = 3.73QKK210 pKa = 11.01IIFPFSKK217 pKa = 10.23SEE219 pKa = 3.83RR220 pKa = 11.84KK221 pKa = 9.4AVGFSFSNQINNRR234 pKa = 11.84AEE236 pKa = 3.92RR237 pKa = 11.84NEE239 pKa = 3.86GGRR242 pKa = 11.84CLLASTAVAKK252 pKa = 8.2DD253 pKa = 3.54TQLEE257 pKa = 4.09GDD259 pKa = 4.41LLNTITRR266 pKa = 11.84KK267 pKa = 8.92FHH269 pKa = 7.38AMPQDD274 pKa = 3.31HH275 pKa = 7.01VNYY278 pKa = 10.24QLFDD282 pKa = 4.94RR283 pKa = 11.84IIRR286 pKa = 11.84EE287 pKa = 4.09ANAILDD293 pKa = 4.16PEE295 pKa = 4.76DD296 pKa = 3.47VDD298 pKa = 4.14AADD301 pKa = 3.93GAGFLSQRR309 pKa = 11.84SNIDD313 pKa = 3.55PLWLTFYY320 pKa = 10.8FAFKK324 pKa = 10.42VIKK327 pKa = 10.54LSVADD332 pKa = 4.51FYY334 pKa = 11.39KK335 pKa = 9.63WQRR338 pKa = 11.84AIYY341 pKa = 7.33ATCRR345 pKa = 11.84KK346 pKa = 9.36IRR348 pKa = 11.84GKK350 pKa = 10.54EE351 pKa = 3.74NIRR354 pKa = 11.84AVLATMLDD362 pKa = 3.67LPGVQDD368 pKa = 3.7TLGKK372 pKa = 9.47MSQSTEE378 pKa = 3.18IHH380 pKa = 5.25TPPWVMHH387 pKa = 6.13FSRR390 pKa = 11.84LYY392 pKa = 10.67DD393 pKa = 3.3IAAVICPCDD402 pKa = 3.78AYY404 pKa = 11.33NVAATIQTPAISWRR418 pKa = 11.84YY419 pKa = 9.69LFTVNPAVKK428 pKa = 10.38GFISYY433 pKa = 7.81LTTLNAVEE441 pKa = 4.27KK442 pKa = 9.45TADD445 pKa = 3.58EE446 pKa = 4.2TSEE449 pKa = 3.66IHH451 pKa = 5.75QSEE454 pKa = 4.6RR455 pKa = 11.84MLQQFILGRR464 pKa = 11.84NSPLANTNHH473 pKa = 6.04LTGNALDD480 pKa = 3.82VTIVGG485 pKa = 4.02
Molecular weight: 53.84 kDa
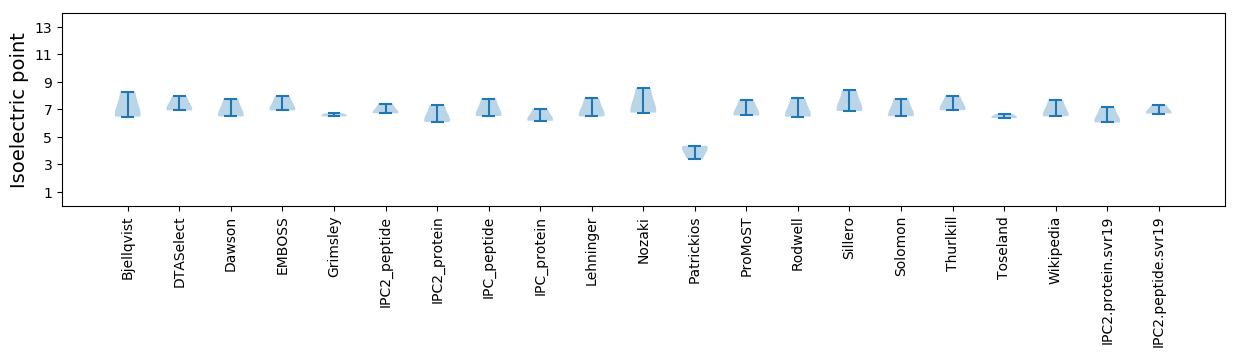
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KS14|A0A0B5KS14_9VIRU Glycoprotein G2 OS=Sanxia Water Strider Virus 1 OX=1608060 GN=G PE=4 SV=1
MM1 pKa = 7.05MLSYY5 pKa = 10.79CATRR9 pKa = 11.84KK10 pKa = 9.76VIPLWTVYY18 pKa = 10.37CYY20 pKa = 10.94LILATVNTVDD30 pKa = 4.64CNLEE34 pKa = 4.03GLSQNDD40 pKa = 3.59LPISDD45 pKa = 4.72RR46 pKa = 11.84LQIVQSTASDD56 pKa = 3.83PSNGALIKK64 pKa = 10.7YY65 pKa = 7.37MYY67 pKa = 9.93HH68 pKa = 6.16GRR70 pKa = 11.84QFDD73 pKa = 4.09TIHH76 pKa = 6.73FNFLMSSSEE85 pKa = 3.99FTLLKK90 pKa = 10.31PKK92 pKa = 10.78NSTCAILKK100 pKa = 10.4GPLIDD105 pKa = 5.47KK106 pKa = 10.54EE107 pKa = 4.26DD108 pKa = 4.02FKK110 pKa = 11.63HH111 pKa = 5.07FTFYY115 pKa = 11.14SATCTIKK122 pKa = 10.34RR123 pKa = 11.84YY124 pKa = 9.87EE125 pKa = 4.48DD126 pKa = 3.41FTVSVVGEE134 pKa = 4.11DD135 pKa = 3.45KK136 pKa = 10.32KK137 pKa = 9.59TSLSFNDD144 pKa = 4.57DD145 pKa = 3.28LSTTCHH151 pKa = 6.68SSLTSVWQFQLTTSNEE167 pKa = 3.94HH168 pKa = 6.46LSINNFLLCKK178 pKa = 10.38NGSLFNIQQSNILIHH193 pKa = 6.68EE194 pKa = 4.43CTTLYY199 pKa = 10.57LIKK202 pKa = 10.56NLQPCLEE209 pKa = 4.71PLSQGKK215 pKa = 8.9KK216 pKa = 8.33VCSISSWKK224 pKa = 10.66SEE226 pKa = 3.89GEE228 pKa = 3.87DD229 pKa = 4.44LKK231 pKa = 11.53LHH233 pKa = 6.64LLNSPSGSINIITDD247 pKa = 3.0TDD249 pKa = 4.03KK250 pKa = 11.01IQRR253 pKa = 11.84FCNSACEE260 pKa = 4.21FKK262 pKa = 11.06VRR264 pKa = 11.84AINYY268 pKa = 9.3ISLTCTDD275 pKa = 3.81GSQDD279 pKa = 3.16FKK281 pKa = 11.14QVHH284 pKa = 5.74KK285 pKa = 10.27EE286 pKa = 4.06VVEE289 pKa = 4.13SCPFITSSKK298 pKa = 9.94IGYY301 pKa = 8.58LALTFCRR308 pKa = 11.84LSTHH312 pKa = 7.13PLLLWFLVLWIILGYY327 pKa = 8.29PICILFSWISTKK339 pKa = 10.52LYY341 pKa = 10.04RR342 pKa = 11.84LSIYY346 pKa = 8.55VFRR349 pKa = 11.84YY350 pKa = 10.09VSIKK354 pKa = 10.47KK355 pKa = 10.08CINHH359 pKa = 6.82NICYY363 pKa = 9.93DD364 pKa = 3.66CNSEE368 pKa = 4.24YY369 pKa = 11.09KK370 pKa = 10.16FDD372 pKa = 4.14NLIEE376 pKa = 4.03LHH378 pKa = 5.42STCINGICPYY388 pKa = 9.93CRR390 pKa = 11.84VGVGEE395 pKa = 4.0NVIDD399 pKa = 3.87HH400 pKa = 6.19VKK402 pKa = 9.33SCKK405 pKa = 10.05SKK407 pKa = 8.68VYY409 pKa = 10.01RR410 pKa = 11.84YY411 pKa = 10.03KK412 pKa = 10.82EE413 pKa = 3.88QLTLLQSNSKK423 pKa = 10.12LEE425 pKa = 4.14HH426 pKa = 5.92GAALTVSIIRR436 pKa = 11.84SKK438 pKa = 10.39LTIITIYY445 pKa = 10.62ILVLLFAMVPMTNSEE460 pKa = 4.11HH461 pKa = 6.98FIPNITDD468 pKa = 3.2QYY470 pKa = 9.11GHH472 pKa = 6.51VIEE475 pKa = 5.4FNTTAEE481 pKa = 4.78DD482 pKa = 3.81YY483 pKa = 11.38LSLFKK488 pKa = 10.84STDD491 pKa = 3.4SLLKK495 pKa = 9.62PAQGNDD501 pKa = 3.71YY502 pKa = 10.1YY503 pKa = 11.72SKK505 pKa = 10.7LASEE509 pKa = 5.4LKK511 pKa = 10.48RR512 pKa = 11.84CDD514 pKa = 3.86FNCFHH519 pKa = 7.03VNNEE523 pKa = 4.27CQCEE527 pKa = 3.99DD528 pKa = 3.35RR529 pKa = 11.84RR530 pKa = 11.84VHH532 pKa = 6.57RR533 pKa = 11.84FKK535 pKa = 11.06RR536 pKa = 11.84DD537 pKa = 3.19LSSNNLGHH545 pKa = 6.11YY546 pKa = 9.45PIVSPKK552 pKa = 10.81GFINYY557 pKa = 9.49NSTYY561 pKa = 10.55QPTSDD566 pKa = 5.24DD567 pKa = 3.42EE568 pKa = 4.76TIQLTFNTITKK579 pKa = 8.93TPTTSVVSGEE589 pKa = 4.34SIYY592 pKa = 11.08RR593 pKa = 11.84MPFQNNEE600 pKa = 4.22GISFRR605 pKa = 11.84ISTDD609 pKa = 2.49EE610 pKa = 4.07DD611 pKa = 4.32FEE613 pKa = 4.86VKK615 pKa = 9.82TISVSLHH622 pKa = 6.84DD623 pKa = 4.4AFQVYY628 pKa = 9.59KK629 pKa = 10.64SKK631 pKa = 10.43FVKK634 pKa = 9.27WVSDD638 pKa = 3.31RR639 pKa = 11.84TLKK642 pKa = 10.47ISEE645 pKa = 4.31EE646 pKa = 4.24FGCTGSCDD654 pKa = 4.05DD655 pKa = 4.44LCKK658 pKa = 10.84CEE660 pKa = 4.87ISTCKK665 pKa = 8.46VAKK668 pKa = 9.14WQDD671 pKa = 3.28DD672 pKa = 3.99RR673 pKa = 11.84GWGCNPSWCWSISDD687 pKa = 4.67GCTCCKK693 pKa = 10.56ASALEE698 pKa = 4.1NFSSGLLGIWEE709 pKa = 4.33VEE711 pKa = 3.6LSYY714 pKa = 11.13FGLTLCLSDD723 pKa = 5.0SMLSYY728 pKa = 9.87KK729 pKa = 10.27CKK731 pKa = 10.01YY732 pKa = 10.51LEE734 pKa = 4.56GPGTFLIGKK743 pKa = 9.07YY744 pKa = 8.6EE745 pKa = 3.9LSIGNPTGNIKK756 pKa = 10.29KK757 pKa = 9.49VSKK760 pKa = 10.64FIATKK765 pKa = 9.99HH766 pKa = 5.55NVGQTHH772 pKa = 6.26SNIDD776 pKa = 3.79QIVEE780 pKa = 4.14IYY782 pKa = 10.4SDD784 pKa = 3.97PNICYY789 pKa = 9.71QEE791 pKa = 4.35HH792 pKa = 6.2CTHH795 pKa = 6.81GEE797 pKa = 3.48IGDD800 pKa = 3.59YY801 pKa = 9.86SFRR804 pKa = 11.84SIEE807 pKa = 4.85DD808 pKa = 2.83ISTYY812 pKa = 10.98ADD814 pKa = 3.66KK815 pKa = 11.22KK816 pKa = 7.86PTTYY820 pKa = 9.91MVWKK824 pKa = 6.25TTSLSRR830 pKa = 11.84QCSFGNYY837 pKa = 6.91PKK839 pKa = 10.43CYY841 pKa = 9.35SDD843 pKa = 5.78DD844 pKa = 3.94MVSSLSDD851 pKa = 3.1KK852 pKa = 10.74FDD854 pKa = 3.4YY855 pKa = 10.16FYY857 pKa = 11.43KK858 pKa = 10.67NKK860 pKa = 10.24QILKK864 pKa = 8.45NDD866 pKa = 3.3FRR868 pKa = 11.84IVTSTLNQDD877 pKa = 3.11RR878 pKa = 11.84SGNPSLVLKK887 pKa = 8.23VQPKK891 pKa = 8.81MNYY894 pKa = 9.84GYY896 pKa = 10.06IEE898 pKa = 3.93GLIKK902 pKa = 10.74VRR904 pKa = 11.84GLEE907 pKa = 4.05LQHH910 pKa = 6.82KK911 pKa = 8.47SVSVKK916 pKa = 9.97VDD918 pKa = 2.87TYY920 pKa = 11.17ILKK923 pKa = 8.91TCGGCRR929 pKa = 11.84NCRR932 pKa = 11.84RR933 pKa = 11.84GFWCTLRR940 pKa = 11.84LSISEE945 pKa = 3.91PLKK948 pKa = 11.04FNIHH952 pKa = 6.91LYY954 pKa = 10.76SEE956 pKa = 5.0DD957 pKa = 3.45PTVAIDD963 pKa = 3.27TRR965 pKa = 11.84TIEE968 pKa = 3.89VGRR971 pKa = 11.84YY972 pKa = 7.28PKK974 pKa = 10.33DD975 pKa = 3.48YY976 pKa = 9.31NVSMFTPILIRR987 pKa = 11.84EE988 pKa = 4.02VKK990 pKa = 9.98ICIKK994 pKa = 10.31EE995 pKa = 3.73VDD997 pKa = 3.32MCKK1000 pKa = 9.9TIKK1003 pKa = 10.57NISLDD1008 pKa = 3.66EE1009 pKa = 4.13PSFYY1013 pKa = 10.12LTHH1016 pKa = 6.79QGDD1019 pKa = 4.41SIYY1022 pKa = 11.16SSLPNKK1028 pKa = 9.94TYY1030 pKa = 10.76DD1031 pKa = 3.7GQNLFYY1037 pKa = 10.65SIYY1040 pKa = 10.61GGIKK1044 pKa = 8.2ATTVAMFDD1052 pKa = 3.72VIRR1055 pKa = 11.84GFWSNIYY1062 pKa = 9.42IVLMVASVLVLLIFLKK1078 pKa = 11.01YY1079 pKa = 10.32LGLLRR1084 pKa = 11.84LVFDD1088 pKa = 3.82VLTCGFCRR1096 pKa = 11.84RR1097 pKa = 11.84KK1098 pKa = 10.13VRR1100 pKa = 11.84YY1101 pKa = 9.22NKK1103 pKa = 10.23SAEE1106 pKa = 4.11KK1107 pKa = 10.52KK1108 pKa = 9.6PLLKK1112 pKa = 9.88RR1113 pKa = 11.84TT1114 pKa = 3.72
MM1 pKa = 7.05MLSYY5 pKa = 10.79CATRR9 pKa = 11.84KK10 pKa = 9.76VIPLWTVYY18 pKa = 10.37CYY20 pKa = 10.94LILATVNTVDD30 pKa = 4.64CNLEE34 pKa = 4.03GLSQNDD40 pKa = 3.59LPISDD45 pKa = 4.72RR46 pKa = 11.84LQIVQSTASDD56 pKa = 3.83PSNGALIKK64 pKa = 10.7YY65 pKa = 7.37MYY67 pKa = 9.93HH68 pKa = 6.16GRR70 pKa = 11.84QFDD73 pKa = 4.09TIHH76 pKa = 6.73FNFLMSSSEE85 pKa = 3.99FTLLKK90 pKa = 10.31PKK92 pKa = 10.78NSTCAILKK100 pKa = 10.4GPLIDD105 pKa = 5.47KK106 pKa = 10.54EE107 pKa = 4.26DD108 pKa = 4.02FKK110 pKa = 11.63HH111 pKa = 5.07FTFYY115 pKa = 11.14SATCTIKK122 pKa = 10.34RR123 pKa = 11.84YY124 pKa = 9.87EE125 pKa = 4.48DD126 pKa = 3.41FTVSVVGEE134 pKa = 4.11DD135 pKa = 3.45KK136 pKa = 10.32KK137 pKa = 9.59TSLSFNDD144 pKa = 4.57DD145 pKa = 3.28LSTTCHH151 pKa = 6.68SSLTSVWQFQLTTSNEE167 pKa = 3.94HH168 pKa = 6.46LSINNFLLCKK178 pKa = 10.38NGSLFNIQQSNILIHH193 pKa = 6.68EE194 pKa = 4.43CTTLYY199 pKa = 10.57LIKK202 pKa = 10.56NLQPCLEE209 pKa = 4.71PLSQGKK215 pKa = 8.9KK216 pKa = 8.33VCSISSWKK224 pKa = 10.66SEE226 pKa = 3.89GEE228 pKa = 3.87DD229 pKa = 4.44LKK231 pKa = 11.53LHH233 pKa = 6.64LLNSPSGSINIITDD247 pKa = 3.0TDD249 pKa = 4.03KK250 pKa = 11.01IQRR253 pKa = 11.84FCNSACEE260 pKa = 4.21FKK262 pKa = 11.06VRR264 pKa = 11.84AINYY268 pKa = 9.3ISLTCTDD275 pKa = 3.81GSQDD279 pKa = 3.16FKK281 pKa = 11.14QVHH284 pKa = 5.74KK285 pKa = 10.27EE286 pKa = 4.06VVEE289 pKa = 4.13SCPFITSSKK298 pKa = 9.94IGYY301 pKa = 8.58LALTFCRR308 pKa = 11.84LSTHH312 pKa = 7.13PLLLWFLVLWIILGYY327 pKa = 8.29PICILFSWISTKK339 pKa = 10.52LYY341 pKa = 10.04RR342 pKa = 11.84LSIYY346 pKa = 8.55VFRR349 pKa = 11.84YY350 pKa = 10.09VSIKK354 pKa = 10.47KK355 pKa = 10.08CINHH359 pKa = 6.82NICYY363 pKa = 9.93DD364 pKa = 3.66CNSEE368 pKa = 4.24YY369 pKa = 11.09KK370 pKa = 10.16FDD372 pKa = 4.14NLIEE376 pKa = 4.03LHH378 pKa = 5.42STCINGICPYY388 pKa = 9.93CRR390 pKa = 11.84VGVGEE395 pKa = 4.0NVIDD399 pKa = 3.87HH400 pKa = 6.19VKK402 pKa = 9.33SCKK405 pKa = 10.05SKK407 pKa = 8.68VYY409 pKa = 10.01RR410 pKa = 11.84YY411 pKa = 10.03KK412 pKa = 10.82EE413 pKa = 3.88QLTLLQSNSKK423 pKa = 10.12LEE425 pKa = 4.14HH426 pKa = 5.92GAALTVSIIRR436 pKa = 11.84SKK438 pKa = 10.39LTIITIYY445 pKa = 10.62ILVLLFAMVPMTNSEE460 pKa = 4.11HH461 pKa = 6.98FIPNITDD468 pKa = 3.2QYY470 pKa = 9.11GHH472 pKa = 6.51VIEE475 pKa = 5.4FNTTAEE481 pKa = 4.78DD482 pKa = 3.81YY483 pKa = 11.38LSLFKK488 pKa = 10.84STDD491 pKa = 3.4SLLKK495 pKa = 9.62PAQGNDD501 pKa = 3.71YY502 pKa = 10.1YY503 pKa = 11.72SKK505 pKa = 10.7LASEE509 pKa = 5.4LKK511 pKa = 10.48RR512 pKa = 11.84CDD514 pKa = 3.86FNCFHH519 pKa = 7.03VNNEE523 pKa = 4.27CQCEE527 pKa = 3.99DD528 pKa = 3.35RR529 pKa = 11.84RR530 pKa = 11.84VHH532 pKa = 6.57RR533 pKa = 11.84FKK535 pKa = 11.06RR536 pKa = 11.84DD537 pKa = 3.19LSSNNLGHH545 pKa = 6.11YY546 pKa = 9.45PIVSPKK552 pKa = 10.81GFINYY557 pKa = 9.49NSTYY561 pKa = 10.55QPTSDD566 pKa = 5.24DD567 pKa = 3.42EE568 pKa = 4.76TIQLTFNTITKK579 pKa = 8.93TPTTSVVSGEE589 pKa = 4.34SIYY592 pKa = 11.08RR593 pKa = 11.84MPFQNNEE600 pKa = 4.22GISFRR605 pKa = 11.84ISTDD609 pKa = 2.49EE610 pKa = 4.07DD611 pKa = 4.32FEE613 pKa = 4.86VKK615 pKa = 9.82TISVSLHH622 pKa = 6.84DD623 pKa = 4.4AFQVYY628 pKa = 9.59KK629 pKa = 10.64SKK631 pKa = 10.43FVKK634 pKa = 9.27WVSDD638 pKa = 3.31RR639 pKa = 11.84TLKK642 pKa = 10.47ISEE645 pKa = 4.31EE646 pKa = 4.24FGCTGSCDD654 pKa = 4.05DD655 pKa = 4.44LCKK658 pKa = 10.84CEE660 pKa = 4.87ISTCKK665 pKa = 8.46VAKK668 pKa = 9.14WQDD671 pKa = 3.28DD672 pKa = 3.99RR673 pKa = 11.84GWGCNPSWCWSISDD687 pKa = 4.67GCTCCKK693 pKa = 10.56ASALEE698 pKa = 4.1NFSSGLLGIWEE709 pKa = 4.33VEE711 pKa = 3.6LSYY714 pKa = 11.13FGLTLCLSDD723 pKa = 5.0SMLSYY728 pKa = 9.87KK729 pKa = 10.27CKK731 pKa = 10.01YY732 pKa = 10.51LEE734 pKa = 4.56GPGTFLIGKK743 pKa = 9.07YY744 pKa = 8.6EE745 pKa = 3.9LSIGNPTGNIKK756 pKa = 10.29KK757 pKa = 9.49VSKK760 pKa = 10.64FIATKK765 pKa = 9.99HH766 pKa = 5.55NVGQTHH772 pKa = 6.26SNIDD776 pKa = 3.79QIVEE780 pKa = 4.14IYY782 pKa = 10.4SDD784 pKa = 3.97PNICYY789 pKa = 9.71QEE791 pKa = 4.35HH792 pKa = 6.2CTHH795 pKa = 6.81GEE797 pKa = 3.48IGDD800 pKa = 3.59YY801 pKa = 9.86SFRR804 pKa = 11.84SIEE807 pKa = 4.85DD808 pKa = 2.83ISTYY812 pKa = 10.98ADD814 pKa = 3.66KK815 pKa = 11.22KK816 pKa = 7.86PTTYY820 pKa = 9.91MVWKK824 pKa = 6.25TTSLSRR830 pKa = 11.84QCSFGNYY837 pKa = 6.91PKK839 pKa = 10.43CYY841 pKa = 9.35SDD843 pKa = 5.78DD844 pKa = 3.94MVSSLSDD851 pKa = 3.1KK852 pKa = 10.74FDD854 pKa = 3.4YY855 pKa = 10.16FYY857 pKa = 11.43KK858 pKa = 10.67NKK860 pKa = 10.24QILKK864 pKa = 8.45NDD866 pKa = 3.3FRR868 pKa = 11.84IVTSTLNQDD877 pKa = 3.11RR878 pKa = 11.84SGNPSLVLKK887 pKa = 8.23VQPKK891 pKa = 8.81MNYY894 pKa = 9.84GYY896 pKa = 10.06IEE898 pKa = 3.93GLIKK902 pKa = 10.74VRR904 pKa = 11.84GLEE907 pKa = 4.05LQHH910 pKa = 6.82KK911 pKa = 8.47SVSVKK916 pKa = 9.97VDD918 pKa = 2.87TYY920 pKa = 11.17ILKK923 pKa = 8.91TCGGCRR929 pKa = 11.84NCRR932 pKa = 11.84RR933 pKa = 11.84GFWCTLRR940 pKa = 11.84LSISEE945 pKa = 3.91PLKK948 pKa = 11.04FNIHH952 pKa = 6.91LYY954 pKa = 10.76SEE956 pKa = 5.0DD957 pKa = 3.45PTVAIDD963 pKa = 3.27TRR965 pKa = 11.84TIEE968 pKa = 3.89VGRR971 pKa = 11.84YY972 pKa = 7.28PKK974 pKa = 10.33DD975 pKa = 3.48YY976 pKa = 9.31NVSMFTPILIRR987 pKa = 11.84EE988 pKa = 4.02VKK990 pKa = 9.98ICIKK994 pKa = 10.31EE995 pKa = 3.73VDD997 pKa = 3.32MCKK1000 pKa = 9.9TIKK1003 pKa = 10.57NISLDD1008 pKa = 3.66EE1009 pKa = 4.13PSFYY1013 pKa = 10.12LTHH1016 pKa = 6.79QGDD1019 pKa = 4.41SIYY1022 pKa = 11.16SSLPNKK1028 pKa = 9.94TYY1030 pKa = 10.76DD1031 pKa = 3.7GQNLFYY1037 pKa = 10.65SIYY1040 pKa = 10.61GGIKK1044 pKa = 8.2ATTVAMFDD1052 pKa = 3.72VIRR1055 pKa = 11.84GFWSNIYY1062 pKa = 9.42IVLMVASVLVLLIFLKK1078 pKa = 11.01YY1079 pKa = 10.32LGLLRR1084 pKa = 11.84LVFDD1088 pKa = 3.82VLTCGFCRR1096 pKa = 11.84RR1097 pKa = 11.84KK1098 pKa = 10.13VRR1100 pKa = 11.84YY1101 pKa = 9.22NKK1103 pKa = 10.23SAEE1106 pKa = 4.11KK1107 pKa = 10.52KK1108 pKa = 9.6PLLKK1112 pKa = 9.88RR1113 pKa = 11.84TT1114 pKa = 3.72
Molecular weight: 127.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5533 |
485 |
3934 |
1844.3 |
210.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.832 ± 1.288 | 2.223 ± 0.793 |
5.476 ± 0.058 | 6.524 ± 0.985 |
4.573 ± 0.179 | 4.301 ± 0.282 |
1.952 ± 0.122 | 8.531 ± 0.622 |
7.808 ± 0.895 | 10.248 ± 0.534 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.458 ± 0.416 | 5.675 ± 0.25 |
2.91 ± 0.3 | 3.56 ± 0.192 |
4.103 ± 0.316 | 10.645 ± 1.048 |
5.765 ± 0.729 | 4.753 ± 0.743 |
0.976 ± 0.121 | 3.687 ± 0.462 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
