
Bat coronavirus HKU9-3
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Nobecovirus; Rousettus bat coronavirus HKU9
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
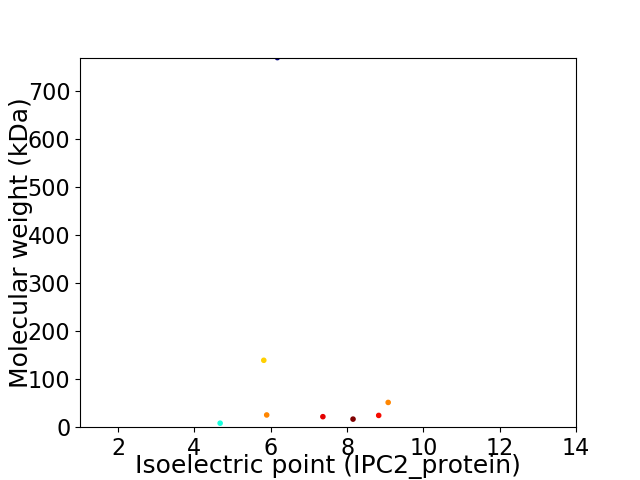
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3EXI5|A3EXI5_BCHK9 Membrane protein OS=Bat coronavirus HKU9-3 OX=424369 GN=M PE=3 SV=1
MM1 pKa = 7.32YY2 pKa = 9.76EE3 pKa = 4.44LVSADD8 pKa = 3.47TSVVIANVLVVIILCLVVVIVGCALLLILQFILGTCGCLFSIICKK53 pKa = 8.67PTILVYY59 pKa = 11.02NKK61 pKa = 9.42FRR63 pKa = 11.84NEE65 pKa = 3.76SLLNEE70 pKa = 4.09QEE72 pKa = 4.04EE73 pKa = 4.36LLII76 pKa = 5.21
MM1 pKa = 7.32YY2 pKa = 9.76EE3 pKa = 4.44LVSADD8 pKa = 3.47TSVVIANVLVVIILCLVVVIVGCALLLILQFILGTCGCLFSIICKK53 pKa = 8.67PTILVYY59 pKa = 11.02NKK61 pKa = 9.42FRR63 pKa = 11.84NEE65 pKa = 3.76SLLNEE70 pKa = 4.09QEE72 pKa = 4.04EE73 pKa = 4.36LLII76 pKa = 5.21
Molecular weight: 8.37 kDa
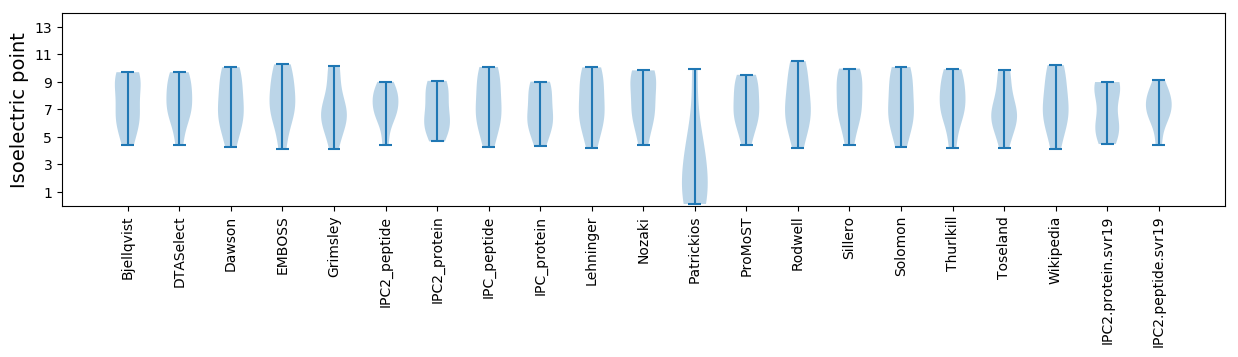
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3EXI7|A3EXI7_BCHK9 Uncharacterized protein NS7a OS=Bat coronavirus HKU9-3 OX=424369 GN=NS7a PE=4 SV=1
MM1 pKa = 7.62SGRR4 pKa = 11.84NKK6 pKa = 9.69PRR8 pKa = 11.84PTSQPKK14 pKa = 8.48VTFKK18 pKa = 10.97QEE20 pKa = 3.89SDD22 pKa = 3.52GSDD25 pKa = 3.37SEE27 pKa = 4.52SEE29 pKa = 3.89RR30 pKa = 11.84RR31 pKa = 11.84NGNRR35 pKa = 11.84SGARR39 pKa = 11.84SKK41 pKa = 11.36NNNGRR46 pKa = 11.84GGAPKK51 pKa = 10.08PEE53 pKa = 4.27KK54 pKa = 10.13PKK56 pKa = 10.56AAPPQNVSWFAPLVQTGKK74 pKa = 10.3QDD76 pKa = 3.27LRR78 pKa = 11.84FPRR81 pKa = 11.84GQGVPISQGVDD92 pKa = 3.34PIYY95 pKa = 11.18NHH97 pKa = 6.99GYY99 pKa = 7.74WVRR102 pKa = 11.84TQRR105 pKa = 11.84TFQKK109 pKa = 10.39GGKK112 pKa = 8.03PVSANPRR119 pKa = 11.84WYY121 pKa = 10.1FYY123 pKa = 9.78YY124 pKa = 10.26TGTGRR129 pKa = 11.84YY130 pKa = 9.52GDD132 pKa = 3.52MRR134 pKa = 11.84YY135 pKa = 8.01GTKK138 pKa = 10.57NPDD141 pKa = 3.26IYY143 pKa = 10.66WVGEE147 pKa = 3.54EE148 pKa = 4.13GANVNRR154 pKa = 11.84VGDD157 pKa = 3.64MGTRR161 pKa = 11.84NPNNDD166 pKa = 3.06AAISVQLADD175 pKa = 5.5GIPKK179 pKa = 10.23GFYY182 pKa = 10.47AEE184 pKa = 4.29GRR186 pKa = 11.84NSRR189 pKa = 11.84GNSRR193 pKa = 11.84NSSRR197 pKa = 11.84NSSRR201 pKa = 11.84ASSRR205 pKa = 11.84GGSRR209 pKa = 11.84PGSRR213 pKa = 11.84GASPGRR219 pKa = 11.84ATPSGSGAEE228 pKa = 3.75PWMAYY233 pKa = 9.69LVSKK237 pKa = 11.29LEE239 pKa = 3.98TLEE242 pKa = 4.54AKK244 pKa = 10.82VNGTKK249 pKa = 10.06SEE251 pKa = 4.19TKK253 pKa = 10.7APVQVTKK260 pKa = 10.85SAAAEE265 pKa = 3.82NAKK268 pKa = 10.28KK269 pKa = 10.1LRR271 pKa = 11.84HH272 pKa = 5.99KK273 pKa = 8.81RR274 pKa = 11.84TPHH277 pKa = 6.14KK278 pKa = 10.85GSGVTMNYY286 pKa = 9.19GRR288 pKa = 11.84RR289 pKa = 11.84GPGDD293 pKa = 3.4LEE295 pKa = 5.04GNFGDD300 pKa = 3.87QTMLKK305 pKa = 10.3LGVDD309 pKa = 4.07DD310 pKa = 4.51PRR312 pKa = 11.84FPAVAQMAPNVASFIFMSHH331 pKa = 6.6LSTRR335 pKa = 11.84EE336 pKa = 3.75EE337 pKa = 4.41NDD339 pKa = 3.98ALWLQYY345 pKa = 10.58KK346 pKa = 10.07GAIKK350 pKa = 10.38LPKK353 pKa = 9.74DD354 pKa = 3.61DD355 pKa = 5.0PNYY358 pKa = 9.59EE359 pKa = 3.76QWTKK363 pKa = 11.25LLAEE367 pKa = 4.58NLNAYY372 pKa = 9.72KK373 pKa = 10.37DD374 pKa = 4.17FPPPEE379 pKa = 4.14PKK381 pKa = 9.44KK382 pKa = 10.51DD383 pKa = 3.35KK384 pKa = 10.53KK385 pKa = 10.65KK386 pKa = 10.72KK387 pKa = 10.23EE388 pKa = 4.24EE389 pKa = 3.97ISSDD393 pKa = 3.52TVVFEE398 pKa = 4.65DD399 pKa = 4.62ASTGTDD405 pKa = 3.19QAVVKK410 pKa = 10.26VWVKK414 pKa = 11.36DD415 pKa = 3.64EE416 pKa = 4.17GAQTDD421 pKa = 4.1DD422 pKa = 3.32EE423 pKa = 4.62WLGGDD428 pKa = 3.25DD429 pKa = 3.35TVYY432 pKa = 10.84EE433 pKa = 4.38EE434 pKa = 4.62EE435 pKa = 4.59DD436 pKa = 4.11DD437 pKa = 3.96RR438 pKa = 11.84PKK440 pKa = 9.38TQRR443 pKa = 11.84RR444 pKa = 11.84HH445 pKa = 5.15KK446 pKa = 10.06KK447 pKa = 9.76RR448 pKa = 11.84NSTASRR454 pKa = 11.84VTIADD459 pKa = 3.43PMNATSEE466 pKa = 4.26RR467 pKa = 11.84SS468 pKa = 3.23
MM1 pKa = 7.62SGRR4 pKa = 11.84NKK6 pKa = 9.69PRR8 pKa = 11.84PTSQPKK14 pKa = 8.48VTFKK18 pKa = 10.97QEE20 pKa = 3.89SDD22 pKa = 3.52GSDD25 pKa = 3.37SEE27 pKa = 4.52SEE29 pKa = 3.89RR30 pKa = 11.84RR31 pKa = 11.84NGNRR35 pKa = 11.84SGARR39 pKa = 11.84SKK41 pKa = 11.36NNNGRR46 pKa = 11.84GGAPKK51 pKa = 10.08PEE53 pKa = 4.27KK54 pKa = 10.13PKK56 pKa = 10.56AAPPQNVSWFAPLVQTGKK74 pKa = 10.3QDD76 pKa = 3.27LRR78 pKa = 11.84FPRR81 pKa = 11.84GQGVPISQGVDD92 pKa = 3.34PIYY95 pKa = 11.18NHH97 pKa = 6.99GYY99 pKa = 7.74WVRR102 pKa = 11.84TQRR105 pKa = 11.84TFQKK109 pKa = 10.39GGKK112 pKa = 8.03PVSANPRR119 pKa = 11.84WYY121 pKa = 10.1FYY123 pKa = 9.78YY124 pKa = 10.26TGTGRR129 pKa = 11.84YY130 pKa = 9.52GDD132 pKa = 3.52MRR134 pKa = 11.84YY135 pKa = 8.01GTKK138 pKa = 10.57NPDD141 pKa = 3.26IYY143 pKa = 10.66WVGEE147 pKa = 3.54EE148 pKa = 4.13GANVNRR154 pKa = 11.84VGDD157 pKa = 3.64MGTRR161 pKa = 11.84NPNNDD166 pKa = 3.06AAISVQLADD175 pKa = 5.5GIPKK179 pKa = 10.23GFYY182 pKa = 10.47AEE184 pKa = 4.29GRR186 pKa = 11.84NSRR189 pKa = 11.84GNSRR193 pKa = 11.84NSSRR197 pKa = 11.84NSSRR201 pKa = 11.84ASSRR205 pKa = 11.84GGSRR209 pKa = 11.84PGSRR213 pKa = 11.84GASPGRR219 pKa = 11.84ATPSGSGAEE228 pKa = 3.75PWMAYY233 pKa = 9.69LVSKK237 pKa = 11.29LEE239 pKa = 3.98TLEE242 pKa = 4.54AKK244 pKa = 10.82VNGTKK249 pKa = 10.06SEE251 pKa = 4.19TKK253 pKa = 10.7APVQVTKK260 pKa = 10.85SAAAEE265 pKa = 3.82NAKK268 pKa = 10.28KK269 pKa = 10.1LRR271 pKa = 11.84HH272 pKa = 5.99KK273 pKa = 8.81RR274 pKa = 11.84TPHH277 pKa = 6.14KK278 pKa = 10.85GSGVTMNYY286 pKa = 9.19GRR288 pKa = 11.84RR289 pKa = 11.84GPGDD293 pKa = 3.4LEE295 pKa = 5.04GNFGDD300 pKa = 3.87QTMLKK305 pKa = 10.3LGVDD309 pKa = 4.07DD310 pKa = 4.51PRR312 pKa = 11.84FPAVAQMAPNVASFIFMSHH331 pKa = 6.6LSTRR335 pKa = 11.84EE336 pKa = 3.75EE337 pKa = 4.41NDD339 pKa = 3.98ALWLQYY345 pKa = 10.58KK346 pKa = 10.07GAIKK350 pKa = 10.38LPKK353 pKa = 9.74DD354 pKa = 3.61DD355 pKa = 5.0PNYY358 pKa = 9.59EE359 pKa = 3.76QWTKK363 pKa = 11.25LLAEE367 pKa = 4.58NLNAYY372 pKa = 9.72KK373 pKa = 10.37DD374 pKa = 4.17FPPPEE379 pKa = 4.14PKK381 pKa = 9.44KK382 pKa = 10.51DD383 pKa = 3.35KK384 pKa = 10.53KK385 pKa = 10.65KK386 pKa = 10.72KK387 pKa = 10.23EE388 pKa = 4.24EE389 pKa = 3.97ISSDD393 pKa = 3.52TVVFEE398 pKa = 4.65DD399 pKa = 4.62ASTGTDD405 pKa = 3.19QAVVKK410 pKa = 10.26VWVKK414 pKa = 11.36DD415 pKa = 3.64EE416 pKa = 4.17GAQTDD421 pKa = 4.1DD422 pKa = 3.32EE423 pKa = 4.62WLGGDD428 pKa = 3.25DD429 pKa = 3.35TVYY432 pKa = 10.84EE433 pKa = 4.38EE434 pKa = 4.62EE435 pKa = 4.59DD436 pKa = 4.11DD437 pKa = 3.96RR438 pKa = 11.84PKK440 pKa = 9.38TQRR443 pKa = 11.84RR444 pKa = 11.84HH445 pKa = 5.15KK446 pKa = 10.06KK447 pKa = 9.76RR448 pKa = 11.84NSTASRR454 pKa = 11.84VTIADD459 pKa = 3.43PMNATSEE466 pKa = 4.26RR467 pKa = 11.84SS468 pKa = 3.23
Molecular weight: 51.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9532 |
76 |
6923 |
1191.5 |
132.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.753 ± 0.209 | 3.063 ± 0.413 |
5.277 ± 0.47 | 3.714 ± 0.564 |
4.144 ± 0.274 | 6.295 ± 0.544 |
1.836 ± 0.168 | 4.091 ± 0.589 |
4.522 ± 0.725 | 9.851 ± 0.754 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.465 ± 0.205 | 4.994 ± 0.792 |
4.658 ± 0.51 | 3.294 ± 0.333 |
4.438 ± 0.696 | 6.756 ± 0.576 |
6.504 ± 0.328 | 9.987 ± 0.709 |
1.311 ± 0.168 | 5.046 ± 0.435 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
