
Pseudomonas sp. ok602
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; unclassified Pseudomonas
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
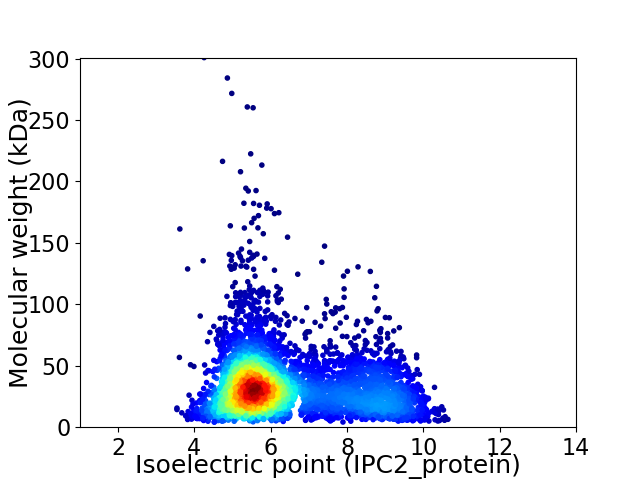
Virtual 2D-PAGE plot for 4728 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4SRV4|A0A1I4SRV4_9PSED Dihydroorotase OS=Pseudomonas sp. ok602 OX=1761898 GN=SAMN04487858_10580 PE=4 SV=1
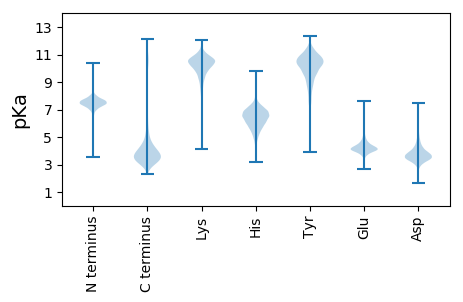
TT1 pKa = 7.12GPITAGSFTGTYY13 pKa = 7.5GTLVLNANGTYY24 pKa = 9.92TYY26 pKa = 10.14TLHH29 pKa = 6.76PNEE32 pKa = 5.26AGFKK36 pKa = 9.95SLPGNGNGIEE46 pKa = 4.29TFTYY50 pKa = 10.35TLTDD54 pKa = 3.32ADD56 pKa = 4.37GDD58 pKa = 3.79QSIANLVLNVHH69 pKa = 6.31NNDD72 pKa = 4.25DD73 pKa = 4.14PVTLGGIKK81 pKa = 10.55LEE83 pKa = 4.5GSDD86 pKa = 3.22LTVYY90 pKa = 10.06EE91 pKa = 4.81KK92 pKa = 10.74NLPMGSDD99 pKa = 3.85PNNGALTQSGTFAVSAPDD117 pKa = 3.5GLKK120 pKa = 9.31TLTVQGITVVDD131 pKa = 3.94GGVTQTIPQSITTALGNKK149 pKa = 8.12LTITGYY155 pKa = 10.29DD156 pKa = 3.03ASTGVVSYY164 pKa = 10.86SYY166 pKa = 9.73TLTGTEE172 pKa = 3.89VHH174 pKa = 6.21PTGLGPNSIVEE185 pKa = 4.46SFTVIASDD193 pKa = 3.85GDD195 pKa = 3.85SGATNTATGTLEE207 pKa = 3.89VNIIDD212 pKa = 5.12DD213 pKa = 4.71LPTAKK218 pKa = 9.84PDD220 pKa = 3.2TGSAIEE226 pKa = 4.24GATLNLSVLGNDD238 pKa = 3.48IKK240 pKa = 11.27GSDD243 pKa = 3.67GAVVIGVRR251 pKa = 11.84GGTNTGTTASGGLNSNISGAYY272 pKa = 10.03GYY274 pKa = 8.01LTLDD278 pKa = 3.27AAGNAVYY285 pKa = 10.18HH286 pKa = 6.15SNPNSVTTGTVTDD299 pKa = 3.6TFVYY303 pKa = 10.25SIRR306 pKa = 11.84DD307 pKa = 3.19GDD309 pKa = 4.14GDD311 pKa = 3.85VSTTTITITINDD323 pKa = 3.79SGLNAVIDD331 pKa = 3.86NDD333 pKa = 3.54VTVYY337 pKa = 10.72EE338 pKa = 4.5KK339 pKa = 11.12ALDD342 pKa = 3.93LKK344 pKa = 11.4ADD346 pKa = 4.11GLDD349 pKa = 3.64LAPGTVIGSDD359 pKa = 3.42PSNTGEE365 pKa = 4.24TAGGTLVGSVTGGSGPLTYY384 pKa = 10.95SLVGSNAGLFGTIQVNADD402 pKa = 2.97GTYY405 pKa = 10.03TYY407 pKa = 10.02TLTKK411 pKa = 10.27AASTSPNANDD421 pKa = 5.0GVTTLNNADD430 pKa = 3.54SFIYY434 pKa = 10.32KK435 pKa = 9.01ATDD438 pKa = 3.07AQGNFVTGTILINIVDD454 pKa = 4.73DD455 pKa = 4.36VPSATASTRR464 pKa = 11.84SVTTVPISTNLMIVLDD480 pKa = 3.95VSGSMKK486 pKa = 10.16EE487 pKa = 3.92LSGVMINGVNQTRR500 pKa = 11.84LALAKK505 pKa = 10.02QAISALLDD513 pKa = 3.55KK514 pKa = 11.55YY515 pKa = 10.89EE516 pKa = 4.16GLGDD520 pKa = 3.8VMVQLVTFSSTAKK533 pKa = 10.49DD534 pKa = 3.54VTTTWVSAAVAKK546 pKa = 10.47AALTALIADD555 pKa = 4.55GGTNYY560 pKa = 10.46DD561 pKa = 3.28AAVAMMQTAFGQTGKK576 pKa = 10.53LAGAQNVGYY585 pKa = 9.35FFSDD589 pKa = 4.12GEE591 pKa = 4.17PSFGQDD597 pKa = 2.61IAPADD602 pKa = 3.76EE603 pKa = 4.61AALKK607 pKa = 9.94TFLTTNAINNYY618 pKa = 10.1AIGLGGGISGGSLDD632 pKa = 3.59PLAYY636 pKa = 10.58NGATGTNTNAVLVTDD651 pKa = 4.77LSQLNSVLSSSVVGVPVTGSLLEE674 pKa = 4.55GGSFGADD681 pKa = 2.53GGFIKK686 pKa = 10.71SIVVDD691 pKa = 4.09GTTYY695 pKa = 10.3TYY697 pKa = 11.1NPKK700 pKa = 9.1LTGSAALTFAGINHH714 pKa = 7.17GLYY717 pKa = 10.47NATDD721 pKa = 3.41HH722 pKa = 6.74SLTITTNNGGTLVIKK737 pKa = 10.42LDD739 pKa = 3.81SGDD742 pKa = 3.57YY743 pKa = 10.77SYY745 pKa = 11.38ISQKK749 pKa = 9.37YY750 pKa = 4.18TTTLITEE757 pKa = 4.19NFGFTLSDD765 pKa = 3.36NDD767 pKa = 3.62GDD769 pKa = 4.4LASSTLTVKK778 pKa = 10.17VLPNTAPVAVDD789 pKa = 2.89DD790 pKa = 4.69HH791 pKa = 7.15VITNTLFSNLTVPGEE806 pKa = 3.93LLLANDD812 pKa = 4.42SDD814 pKa = 4.69PNGDD818 pKa = 3.57RR819 pKa = 11.84LTSNPANFNTGSDD832 pKa = 3.7FRR834 pKa = 11.84GSGAITTFALPDD846 pKa = 3.48NLAANQNLADD856 pKa = 3.76VRR858 pKa = 11.84NAFAANVATMTAVLVISGYY877 pKa = 10.39LGEE880 pKa = 4.33VSGANANDD888 pKa = 3.49EE889 pKa = 4.6DD890 pKa = 4.77FISVKK895 pKa = 10.51LKK897 pKa = 10.83AGEE900 pKa = 4.23TLTLDD905 pKa = 3.29HH906 pKa = 6.72NLAGGHH912 pKa = 5.37VSMEE916 pKa = 3.93YY917 pKa = 10.6SLNGAGFLPISDD929 pKa = 4.05GGMITALTDD938 pKa = 2.95GTYY941 pKa = 10.15QIHH944 pKa = 5.44VTNITNTNGNNKK956 pKa = 8.87GAAEE960 pKa = 4.32SYY962 pKa = 9.88QLTMTVDD969 pKa = 3.38YY970 pKa = 10.57GAVHH974 pKa = 6.61GTTSDD979 pKa = 3.32YY980 pKa = 11.19HH981 pKa = 5.18GTYY984 pKa = 8.61TASDD988 pKa = 3.38NHH990 pKa = 6.58GGTDD994 pKa = 3.72GANVTLTYY1002 pKa = 10.4QAGHH1006 pKa = 6.42TLTGTSGDD1014 pKa = 4.55DD1015 pKa = 3.12ILVASGDD1022 pKa = 3.82NNLLNGGDD1030 pKa = 4.0GNDD1033 pKa = 3.23VLTVSGINNEE1043 pKa = 4.1LHH1045 pKa = 6.65GGAGNDD1051 pKa = 4.09LLFSGPGADD1060 pKa = 4.88LLDD1063 pKa = 4.41GGTGIDD1069 pKa = 3.47TASYY1073 pKa = 9.24AHH1075 pKa = 6.58ATAGVTVNLGTLIAQNTIGAGTDD1098 pKa = 3.13TLTSIEE1104 pKa = 4.21NLVGSNYY1111 pKa = 10.14NDD1113 pKa = 3.6TLTGDD1118 pKa = 3.39NGNNVINGGLGNDD1131 pKa = 3.86TLNGGGGDD1139 pKa = 4.55DD1140 pKa = 3.95ILIGGLGNNTLSGGAGADD1158 pKa = 3.36TFLWQQGNSGHH1169 pKa = 7.16DD1170 pKa = 3.38VVTDD1174 pKa = 3.95FTLGTDD1180 pKa = 4.27KK1181 pKa = 11.37LDD1183 pKa = 4.71LSQLLQGEE1191 pKa = 4.5NATTASLDD1199 pKa = 3.81DD1200 pKa = 3.89YY1201 pKa = 11.27LHH1203 pKa = 6.67FSVTVVGGAVVSSITVSSIAGAAANQTIDD1232 pKa = 3.48LAGVNLADD1240 pKa = 4.46HH1241 pKa = 6.53YY1242 pKa = 11.4GVTPGAGGAIASGHH1256 pKa = 5.79DD1257 pKa = 3.17TATIITGMLNDD1268 pKa = 4.5HH1269 pKa = 6.1SLKK1272 pKa = 10.51VDD1274 pKa = 3.57TVV1276 pKa = 3.29
TT1 pKa = 7.12GPITAGSFTGTYY13 pKa = 7.5GTLVLNANGTYY24 pKa = 9.92TYY26 pKa = 10.14TLHH29 pKa = 6.76PNEE32 pKa = 5.26AGFKK36 pKa = 9.95SLPGNGNGIEE46 pKa = 4.29TFTYY50 pKa = 10.35TLTDD54 pKa = 3.32ADD56 pKa = 4.37GDD58 pKa = 3.79QSIANLVLNVHH69 pKa = 6.31NNDD72 pKa = 4.25DD73 pKa = 4.14PVTLGGIKK81 pKa = 10.55LEE83 pKa = 4.5GSDD86 pKa = 3.22LTVYY90 pKa = 10.06EE91 pKa = 4.81KK92 pKa = 10.74NLPMGSDD99 pKa = 3.85PNNGALTQSGTFAVSAPDD117 pKa = 3.5GLKK120 pKa = 9.31TLTVQGITVVDD131 pKa = 3.94GGVTQTIPQSITTALGNKK149 pKa = 8.12LTITGYY155 pKa = 10.29DD156 pKa = 3.03ASTGVVSYY164 pKa = 10.86SYY166 pKa = 9.73TLTGTEE172 pKa = 3.89VHH174 pKa = 6.21PTGLGPNSIVEE185 pKa = 4.46SFTVIASDD193 pKa = 3.85GDD195 pKa = 3.85SGATNTATGTLEE207 pKa = 3.89VNIIDD212 pKa = 5.12DD213 pKa = 4.71LPTAKK218 pKa = 9.84PDD220 pKa = 3.2TGSAIEE226 pKa = 4.24GATLNLSVLGNDD238 pKa = 3.48IKK240 pKa = 11.27GSDD243 pKa = 3.67GAVVIGVRR251 pKa = 11.84GGTNTGTTASGGLNSNISGAYY272 pKa = 10.03GYY274 pKa = 8.01LTLDD278 pKa = 3.27AAGNAVYY285 pKa = 10.18HH286 pKa = 6.15SNPNSVTTGTVTDD299 pKa = 3.6TFVYY303 pKa = 10.25SIRR306 pKa = 11.84DD307 pKa = 3.19GDD309 pKa = 4.14GDD311 pKa = 3.85VSTTTITITINDD323 pKa = 3.79SGLNAVIDD331 pKa = 3.86NDD333 pKa = 3.54VTVYY337 pKa = 10.72EE338 pKa = 4.5KK339 pKa = 11.12ALDD342 pKa = 3.93LKK344 pKa = 11.4ADD346 pKa = 4.11GLDD349 pKa = 3.64LAPGTVIGSDD359 pKa = 3.42PSNTGEE365 pKa = 4.24TAGGTLVGSVTGGSGPLTYY384 pKa = 10.95SLVGSNAGLFGTIQVNADD402 pKa = 2.97GTYY405 pKa = 10.03TYY407 pKa = 10.02TLTKK411 pKa = 10.27AASTSPNANDD421 pKa = 5.0GVTTLNNADD430 pKa = 3.54SFIYY434 pKa = 10.32KK435 pKa = 9.01ATDD438 pKa = 3.07AQGNFVTGTILINIVDD454 pKa = 4.73DD455 pKa = 4.36VPSATASTRR464 pKa = 11.84SVTTVPISTNLMIVLDD480 pKa = 3.95VSGSMKK486 pKa = 10.16EE487 pKa = 3.92LSGVMINGVNQTRR500 pKa = 11.84LALAKK505 pKa = 10.02QAISALLDD513 pKa = 3.55KK514 pKa = 11.55YY515 pKa = 10.89EE516 pKa = 4.16GLGDD520 pKa = 3.8VMVQLVTFSSTAKK533 pKa = 10.49DD534 pKa = 3.54VTTTWVSAAVAKK546 pKa = 10.47AALTALIADD555 pKa = 4.55GGTNYY560 pKa = 10.46DD561 pKa = 3.28AAVAMMQTAFGQTGKK576 pKa = 10.53LAGAQNVGYY585 pKa = 9.35FFSDD589 pKa = 4.12GEE591 pKa = 4.17PSFGQDD597 pKa = 2.61IAPADD602 pKa = 3.76EE603 pKa = 4.61AALKK607 pKa = 9.94TFLTTNAINNYY618 pKa = 10.1AIGLGGGISGGSLDD632 pKa = 3.59PLAYY636 pKa = 10.58NGATGTNTNAVLVTDD651 pKa = 4.77LSQLNSVLSSSVVGVPVTGSLLEE674 pKa = 4.55GGSFGADD681 pKa = 2.53GGFIKK686 pKa = 10.71SIVVDD691 pKa = 4.09GTTYY695 pKa = 10.3TYY697 pKa = 11.1NPKK700 pKa = 9.1LTGSAALTFAGINHH714 pKa = 7.17GLYY717 pKa = 10.47NATDD721 pKa = 3.41HH722 pKa = 6.74SLTITTNNGGTLVIKK737 pKa = 10.42LDD739 pKa = 3.81SGDD742 pKa = 3.57YY743 pKa = 10.77SYY745 pKa = 11.38ISQKK749 pKa = 9.37YY750 pKa = 4.18TTTLITEE757 pKa = 4.19NFGFTLSDD765 pKa = 3.36NDD767 pKa = 3.62GDD769 pKa = 4.4LASSTLTVKK778 pKa = 10.17VLPNTAPVAVDD789 pKa = 2.89DD790 pKa = 4.69HH791 pKa = 7.15VITNTLFSNLTVPGEE806 pKa = 3.93LLLANDD812 pKa = 4.42SDD814 pKa = 4.69PNGDD818 pKa = 3.57RR819 pKa = 11.84LTSNPANFNTGSDD832 pKa = 3.7FRR834 pKa = 11.84GSGAITTFALPDD846 pKa = 3.48NLAANQNLADD856 pKa = 3.76VRR858 pKa = 11.84NAFAANVATMTAVLVISGYY877 pKa = 10.39LGEE880 pKa = 4.33VSGANANDD888 pKa = 3.49EE889 pKa = 4.6DD890 pKa = 4.77FISVKK895 pKa = 10.51LKK897 pKa = 10.83AGEE900 pKa = 4.23TLTLDD905 pKa = 3.29HH906 pKa = 6.72NLAGGHH912 pKa = 5.37VSMEE916 pKa = 3.93YY917 pKa = 10.6SLNGAGFLPISDD929 pKa = 4.05GGMITALTDD938 pKa = 2.95GTYY941 pKa = 10.15QIHH944 pKa = 5.44VTNITNTNGNNKK956 pKa = 8.87GAAEE960 pKa = 4.32SYY962 pKa = 9.88QLTMTVDD969 pKa = 3.38YY970 pKa = 10.57GAVHH974 pKa = 6.61GTTSDD979 pKa = 3.32YY980 pKa = 11.19HH981 pKa = 5.18GTYY984 pKa = 8.61TASDD988 pKa = 3.38NHH990 pKa = 6.58GGTDD994 pKa = 3.72GANVTLTYY1002 pKa = 10.4QAGHH1006 pKa = 6.42TLTGTSGDD1014 pKa = 4.55DD1015 pKa = 3.12ILVASGDD1022 pKa = 3.82NNLLNGGDD1030 pKa = 4.0GNDD1033 pKa = 3.23VLTVSGINNEE1043 pKa = 4.1LHH1045 pKa = 6.65GGAGNDD1051 pKa = 4.09LLFSGPGADD1060 pKa = 4.88LLDD1063 pKa = 4.41GGTGIDD1069 pKa = 3.47TASYY1073 pKa = 9.24AHH1075 pKa = 6.58ATAGVTVNLGTLIAQNTIGAGTDD1098 pKa = 3.13TLTSIEE1104 pKa = 4.21NLVGSNYY1111 pKa = 10.14NDD1113 pKa = 3.6TLTGDD1118 pKa = 3.39NGNNVINGGLGNDD1131 pKa = 3.86TLNGGGGDD1139 pKa = 4.55DD1140 pKa = 3.95ILIGGLGNNTLSGGAGADD1158 pKa = 3.36TFLWQQGNSGHH1169 pKa = 7.16DD1170 pKa = 3.38VVTDD1174 pKa = 3.95FTLGTDD1180 pKa = 4.27KK1181 pKa = 11.37LDD1183 pKa = 4.71LSQLLQGEE1191 pKa = 4.5NATTASLDD1199 pKa = 3.81DD1200 pKa = 3.89YY1201 pKa = 11.27LHH1203 pKa = 6.67FSVTVVGGAVVSSITVSSIAGAAANQTIDD1232 pKa = 3.48LAGVNLADD1240 pKa = 4.46HH1241 pKa = 6.53YY1242 pKa = 11.4GVTPGAGGAIASGHH1256 pKa = 5.79DD1257 pKa = 3.17TATIITGMLNDD1268 pKa = 4.5HH1269 pKa = 6.1SLKK1272 pKa = 10.51VDD1274 pKa = 3.57TVV1276 pKa = 3.29
Molecular weight: 128.78 kDa
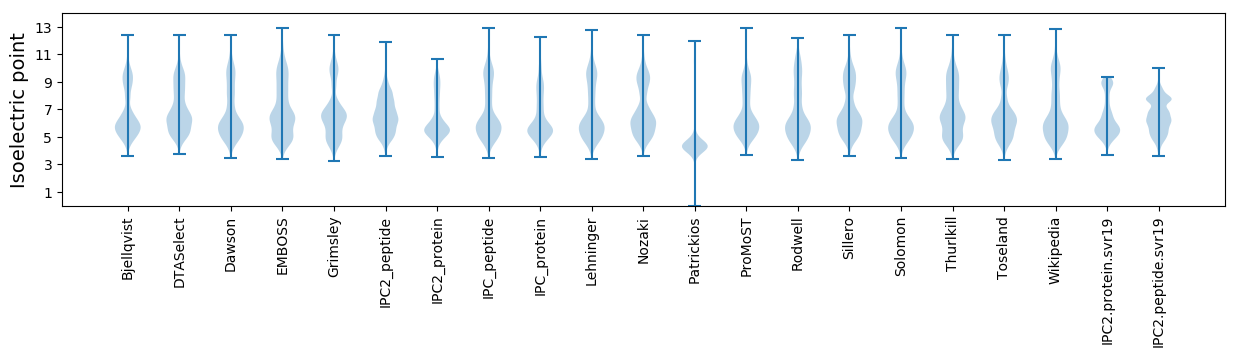
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4WF25|A0A1I4WF25_9PSED Branched-chain alpha-keto acid dehydrogenase E1 component OS=Pseudomonas sp. ok602 OX=1761898 GN=SAMN04487858_112149 PE=4 SV=1
MM1 pKa = 7.55LLASIFGVVMGVVLGLTGAGGGILAVPALVLGLGWNMTQAAPVALFAVGSAAAVGALDD59 pKa = 3.81GLRR62 pKa = 11.84HH63 pKa = 5.17GLVRR67 pKa = 11.84YY68 pKa = 9.27RR69 pKa = 11.84AALLIAVLGALFSPLGIYY87 pKa = 9.11VAHH90 pKa = 6.2QLPDD94 pKa = 3.63NVLMILFSALMVLVAWRR111 pKa = 11.84MLRR114 pKa = 11.84RR115 pKa = 11.84EE116 pKa = 4.02QPAPGPSDD124 pKa = 3.38HH125 pKa = 6.83GQANWGQKK133 pKa = 9.73NCMLDD138 pKa = 3.29QQTGRR143 pKa = 11.84FSWTAKK149 pKa = 8.94CTATLAGLGAVTGVVSGLLGVGGGFLIVPAFKK181 pKa = 10.63QLTDD185 pKa = 3.22VQMRR189 pKa = 11.84GIVATSLMVISLISAIGVLGAFHH212 pKa = 7.06AGVRR216 pKa = 11.84IDD218 pKa = 3.62SQGVAFIVASILGMIIGRR236 pKa = 11.84RR237 pKa = 11.84LCARR241 pKa = 11.84VPARR245 pKa = 11.84ALQMGFASVCLVVAGYY261 pKa = 8.7MLLKK265 pKa = 10.73AA266 pKa = 4.78
MM1 pKa = 7.55LLASIFGVVMGVVLGLTGAGGGILAVPALVLGLGWNMTQAAPVALFAVGSAAAVGALDD59 pKa = 3.81GLRR62 pKa = 11.84HH63 pKa = 5.17GLVRR67 pKa = 11.84YY68 pKa = 9.27RR69 pKa = 11.84AALLIAVLGALFSPLGIYY87 pKa = 9.11VAHH90 pKa = 6.2QLPDD94 pKa = 3.63NVLMILFSALMVLVAWRR111 pKa = 11.84MLRR114 pKa = 11.84RR115 pKa = 11.84EE116 pKa = 4.02QPAPGPSDD124 pKa = 3.38HH125 pKa = 6.83GQANWGQKK133 pKa = 9.73NCMLDD138 pKa = 3.29QQTGRR143 pKa = 11.84FSWTAKK149 pKa = 8.94CTATLAGLGAVTGVVSGLLGVGGGFLIVPAFKK181 pKa = 10.63QLTDD185 pKa = 3.22VQMRR189 pKa = 11.84GIVATSLMVISLISAIGVLGAFHH212 pKa = 7.06AGVRR216 pKa = 11.84IDD218 pKa = 3.62SQGVAFIVASILGMIIGRR236 pKa = 11.84RR237 pKa = 11.84LCARR241 pKa = 11.84VPARR245 pKa = 11.84ALQMGFASVCLVVAGYY261 pKa = 8.7MLLKK265 pKa = 10.73AA266 pKa = 4.78
Molecular weight: 27.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1540914 |
39 |
3116 |
325.9 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.884 ± 0.039 | 0.993 ± 0.01 |
5.378 ± 0.027 | 5.503 ± 0.036 |
3.634 ± 0.019 | 7.886 ± 0.038 |
2.352 ± 0.019 | 4.859 ± 0.027 |
3.573 ± 0.03 | 11.77 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.357 ± 0.017 | 3.098 ± 0.03 |
4.908 ± 0.026 | 4.613 ± 0.026 |
6.219 ± 0.035 | 5.79 ± 0.026 |
5.012 ± 0.034 | 7.163 ± 0.032 |
1.433 ± 0.016 | 2.572 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
