
TM7 phylum sp. oral taxon 348
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Saccharibacteria
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
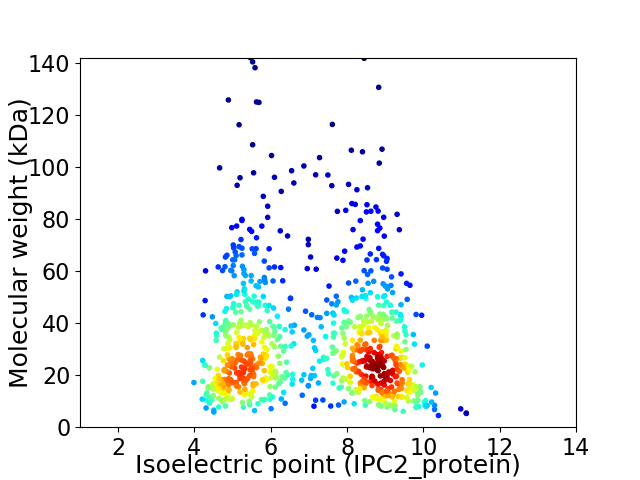
Virtual 2D-PAGE plot for 855 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A563CR36|A0A563CR36_9BACT Uncharacterized protein OS=TM7 phylum sp. oral taxon 348 OX=671231 GN=EUA62_01715 PE=4 SV=1
MM1 pKa = 6.77STSSIYY7 pKa = 9.26YY8 pKa = 7.82TATDD12 pKa = 3.08VPMRR16 pKa = 11.84NEE18 pKa = 3.62SWHH21 pKa = 6.63AEE23 pKa = 4.01QAFKK27 pKa = 10.71SYY29 pKa = 11.33KK30 pKa = 10.36LGGEE34 pKa = 4.04LWFSCVLYY42 pKa = 10.43IVTACVGFVGLFAIDD57 pKa = 5.17SSDD60 pKa = 3.48CPNIVLEE67 pKa = 4.5IFAKK71 pKa = 10.17IYY73 pKa = 8.94FASMCMLGIGYY84 pKa = 9.2LAFAAKK90 pKa = 10.18RR91 pKa = 11.84LMYY94 pKa = 9.99RR95 pKa = 11.84KK96 pKa = 9.81YY97 pKa = 9.9YY98 pKa = 10.36SKK100 pKa = 10.41PSRR103 pKa = 11.84GKK105 pKa = 10.4GEE107 pKa = 4.58LYY109 pKa = 10.24RR110 pKa = 11.84IDD112 pKa = 4.15IIEE115 pKa = 4.9PNDD118 pKa = 4.42DD119 pKa = 3.15DD120 pKa = 4.11WGIQFTSPNGVFTYY134 pKa = 11.04LNLGCDD140 pKa = 4.75LEE142 pKa = 4.9CSMYY146 pKa = 10.65AIEE149 pKa = 5.56LLQFDD154 pKa = 4.1ADD156 pKa = 3.94FRR158 pKa = 11.84SEE160 pKa = 4.08INSYY164 pKa = 10.54LADD167 pKa = 3.86PVNKK171 pKa = 9.87KK172 pKa = 10.21VSTEE176 pKa = 3.54EE177 pKa = 4.15MINSKK182 pKa = 11.02YY183 pKa = 10.74EE184 pKa = 4.01DD185 pKa = 3.84LCDD188 pKa = 4.2DD189 pKa = 4.49RR190 pKa = 11.84PSEE193 pKa = 5.9DD194 pKa = 3.71EE195 pKa = 5.26DD196 pKa = 4.36EE197 pKa = 5.15DD198 pKa = 3.91DD199 pKa = 5.34GRR201 pKa = 11.84YY202 pKa = 9.91VDD204 pKa = 5.44DD205 pKa = 5.1KK206 pKa = 11.42FVV208 pKa = 2.96
MM1 pKa = 6.77STSSIYY7 pKa = 9.26YY8 pKa = 7.82TATDD12 pKa = 3.08VPMRR16 pKa = 11.84NEE18 pKa = 3.62SWHH21 pKa = 6.63AEE23 pKa = 4.01QAFKK27 pKa = 10.71SYY29 pKa = 11.33KK30 pKa = 10.36LGGEE34 pKa = 4.04LWFSCVLYY42 pKa = 10.43IVTACVGFVGLFAIDD57 pKa = 5.17SSDD60 pKa = 3.48CPNIVLEE67 pKa = 4.5IFAKK71 pKa = 10.17IYY73 pKa = 8.94FASMCMLGIGYY84 pKa = 9.2LAFAAKK90 pKa = 10.18RR91 pKa = 11.84LMYY94 pKa = 9.99RR95 pKa = 11.84KK96 pKa = 9.81YY97 pKa = 9.9YY98 pKa = 10.36SKK100 pKa = 10.41PSRR103 pKa = 11.84GKK105 pKa = 10.4GEE107 pKa = 4.58LYY109 pKa = 10.24RR110 pKa = 11.84IDD112 pKa = 4.15IIEE115 pKa = 4.9PNDD118 pKa = 4.42DD119 pKa = 3.15DD120 pKa = 4.11WGIQFTSPNGVFTYY134 pKa = 11.04LNLGCDD140 pKa = 4.75LEE142 pKa = 4.9CSMYY146 pKa = 10.65AIEE149 pKa = 5.56LLQFDD154 pKa = 4.1ADD156 pKa = 3.94FRR158 pKa = 11.84SEE160 pKa = 4.08INSYY164 pKa = 10.54LADD167 pKa = 3.86PVNKK171 pKa = 9.87KK172 pKa = 10.21VSTEE176 pKa = 3.54EE177 pKa = 4.15MINSKK182 pKa = 11.02YY183 pKa = 10.74EE184 pKa = 4.01DD185 pKa = 3.84LCDD188 pKa = 4.2DD189 pKa = 4.49RR190 pKa = 11.84PSEE193 pKa = 5.9DD194 pKa = 3.71EE195 pKa = 5.26DD196 pKa = 4.36EE197 pKa = 5.15DD198 pKa = 3.91DD199 pKa = 5.34GRR201 pKa = 11.84YY202 pKa = 9.91VDD204 pKa = 5.44DD205 pKa = 5.1KK206 pKa = 11.42FVV208 pKa = 2.96
Molecular weight: 23.81 kDa
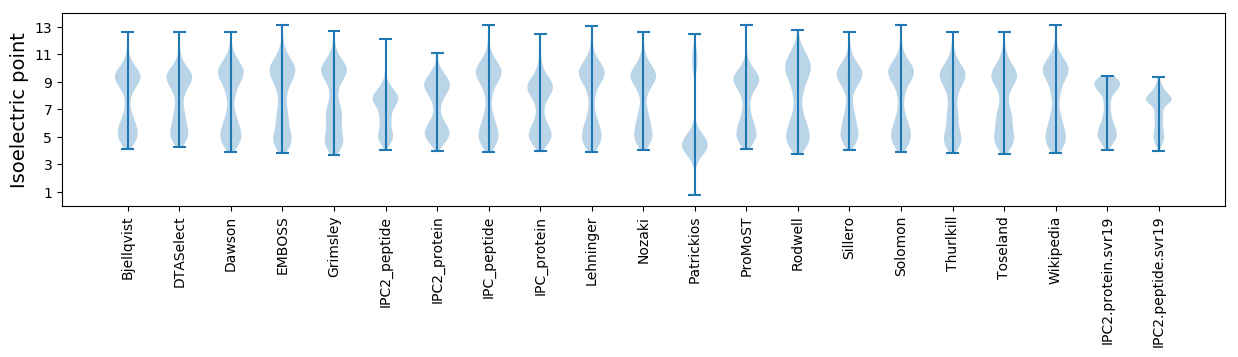
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A563CML3|A0A563CML3_9BACT Transcriptional repressor NrdR OS=TM7 phylum sp. oral taxon 348 OX=671231 GN=nrdR PE=3 SV=1
MM1 pKa = 8.03PKK3 pKa = 8.98RR4 pKa = 11.84TYY6 pKa = 10.09QPHH9 pKa = 5.49KK10 pKa = 9.24RR11 pKa = 11.84SRR13 pKa = 11.84AKK15 pKa = 10.04VHH17 pKa = 5.81GFRR20 pKa = 11.84ARR22 pKa = 11.84MSSRR26 pKa = 11.84AGRR29 pKa = 11.84LVLKK33 pKa = 10.41RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 9.64KK39 pKa = 8.43RR40 pKa = 11.84AKK42 pKa = 9.93IAII45 pKa = 4.09
MM1 pKa = 8.03PKK3 pKa = 8.98RR4 pKa = 11.84TYY6 pKa = 10.09QPHH9 pKa = 5.49KK10 pKa = 9.24RR11 pKa = 11.84SRR13 pKa = 11.84AKK15 pKa = 10.04VHH17 pKa = 5.81GFRR20 pKa = 11.84ARR22 pKa = 11.84MSSRR26 pKa = 11.84AGRR29 pKa = 11.84LVLKK33 pKa = 10.41RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 9.64KK39 pKa = 8.43RR40 pKa = 11.84AKK42 pKa = 9.93IAII45 pKa = 4.09
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
263325 |
38 |
1286 |
308.0 |
34.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.522 ± 0.086 | 0.697 ± 0.024 |
5.968 ± 0.077 | 5.941 ± 0.082 |
3.799 ± 0.063 | 6.459 ± 0.071 |
1.814 ± 0.035 | 8.192 ± 0.091 |
7.104 ± 0.083 | 9.131 ± 0.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.305 ± 0.034 | 5.261 ± 0.073 |
3.713 ± 0.07 | 3.671 ± 0.055 |
5.124 ± 0.069 | 7.068 ± 0.091 |
5.259 ± 0.056 | 6.521 ± 0.069 |
0.96 ± 0.032 | 3.491 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
