
Galbibacter marinus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Galbibacter
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
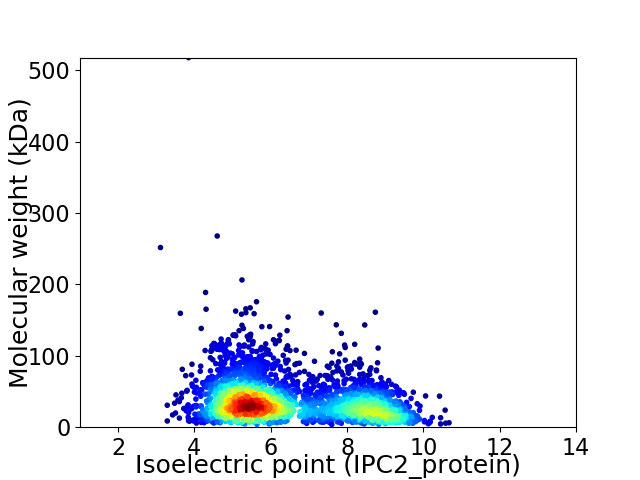
Virtual 2D-PAGE plot for 3099 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K2PV86|K2PV86_9FLAO Glyceraldehyde-3-phosphate dehydrogenase OS=Galbibacter marinus OX=555500 GN=I215_08031 PE=3 SV=1
MM1 pKa = 7.55GGLLISCDD9 pKa = 3.4DD10 pKa = 4.39HH11 pKa = 6.59EE12 pKa = 5.56NSMGEE17 pKa = 4.01KK18 pKa = 10.06LDD20 pKa = 3.82PSEE23 pKa = 4.53LKK25 pKa = 10.85FEE27 pKa = 4.34IEE29 pKa = 3.57QDD31 pKa = 3.47LTIDD35 pKa = 3.28QGGNTVILTNNTPGTIPIWNYY56 pKa = 8.5GTGRR60 pKa = 11.84SNKK63 pKa = 10.22AIDD66 pKa = 3.77TIRR69 pKa = 11.84FAFAGEE75 pKa = 3.88YY76 pKa = 9.6TIEE79 pKa = 4.13FSVMTEE85 pKa = 3.95GGLVEE90 pKa = 5.05ADD92 pKa = 3.41PVTISVTDD100 pKa = 3.41NNFEE104 pKa = 4.24YY105 pKa = 11.13VEE107 pKa = 4.12NEE109 pKa = 3.42LWLKK113 pKa = 10.96LSGGVGNAKK122 pKa = 8.51TWVLDD127 pKa = 3.73LDD129 pKa = 3.99AEE131 pKa = 4.75GVSKK135 pKa = 10.91YY136 pKa = 9.84FVGPQYY142 pKa = 10.76FYY144 pKa = 10.73GTNNGWLEE152 pKa = 4.72GGDD155 pKa = 3.56EE156 pKa = 4.12WSGGSTGCYY165 pKa = 9.99GADD168 pKa = 2.95CWNWNPDD175 pKa = 2.85WPGNSWIMAAGDD187 pKa = 3.93YY188 pKa = 10.18GTMTFSLDD196 pKa = 2.88GGPFVEE202 pKa = 4.92VDD204 pKa = 3.22NLMFAQQGKK213 pKa = 9.63QSGTYY218 pKa = 9.3YY219 pKa = 11.03LDD221 pKa = 4.37ANSHH225 pKa = 5.33TLTMTDD231 pKa = 3.12AQLLHH236 pKa = 6.36NAEE239 pKa = 4.42NDD241 pKa = 3.64DD242 pKa = 4.87CVSNWSDD249 pKa = 3.07MKK251 pKa = 10.79IFSLTEE257 pKa = 3.49DD258 pKa = 3.49TMQLGVLRR266 pKa = 11.84KK267 pKa = 9.7SSCDD271 pKa = 3.01GAAYY275 pKa = 10.33LVFNFISKK283 pKa = 9.91EE284 pKa = 3.95YY285 pKa = 9.64SDD287 pKa = 3.34NWVPEE292 pKa = 4.16EE293 pKa = 4.23TEE295 pKa = 3.87PTIDD299 pKa = 3.71EE300 pKa = 4.68GFDD303 pKa = 3.43PSLEE307 pKa = 4.14AGQLLEE313 pKa = 4.62MLTGGQGSGRR323 pKa = 11.84VWAMDD328 pKa = 3.59AAGNPVDD335 pKa = 4.21WVTAGAGWTVDD346 pKa = 3.43ASSSADD352 pKa = 3.34WGWNSSWSTVASDD365 pKa = 2.58SWIRR369 pKa = 11.84FDD371 pKa = 4.27QWNGMQTYY379 pKa = 8.35TRR381 pKa = 11.84NQSGVEE387 pKa = 3.88TSGTFTINEE396 pKa = 4.09ATNEE400 pKa = 3.88ITLSGEE406 pKa = 4.17VLIQNPGNWMTPTTNTITVIKK427 pKa = 10.62AFDD430 pKa = 4.1DD431 pKa = 4.17YY432 pKa = 9.77QTKK435 pKa = 10.68GIWFGTSYY443 pKa = 11.25DD444 pKa = 3.84SEE446 pKa = 4.63KK447 pKa = 11.28DD448 pKa = 2.84EE449 pKa = 4.38WLAFHH454 pKa = 6.7YY455 pKa = 10.07IIPP458 pKa = 4.18
MM1 pKa = 7.55GGLLISCDD9 pKa = 3.4DD10 pKa = 4.39HH11 pKa = 6.59EE12 pKa = 5.56NSMGEE17 pKa = 4.01KK18 pKa = 10.06LDD20 pKa = 3.82PSEE23 pKa = 4.53LKK25 pKa = 10.85FEE27 pKa = 4.34IEE29 pKa = 3.57QDD31 pKa = 3.47LTIDD35 pKa = 3.28QGGNTVILTNNTPGTIPIWNYY56 pKa = 8.5GTGRR60 pKa = 11.84SNKK63 pKa = 10.22AIDD66 pKa = 3.77TIRR69 pKa = 11.84FAFAGEE75 pKa = 3.88YY76 pKa = 9.6TIEE79 pKa = 4.13FSVMTEE85 pKa = 3.95GGLVEE90 pKa = 5.05ADD92 pKa = 3.41PVTISVTDD100 pKa = 3.41NNFEE104 pKa = 4.24YY105 pKa = 11.13VEE107 pKa = 4.12NEE109 pKa = 3.42LWLKK113 pKa = 10.96LSGGVGNAKK122 pKa = 8.51TWVLDD127 pKa = 3.73LDD129 pKa = 3.99AEE131 pKa = 4.75GVSKK135 pKa = 10.91YY136 pKa = 9.84FVGPQYY142 pKa = 10.76FYY144 pKa = 10.73GTNNGWLEE152 pKa = 4.72GGDD155 pKa = 3.56EE156 pKa = 4.12WSGGSTGCYY165 pKa = 9.99GADD168 pKa = 2.95CWNWNPDD175 pKa = 2.85WPGNSWIMAAGDD187 pKa = 3.93YY188 pKa = 10.18GTMTFSLDD196 pKa = 2.88GGPFVEE202 pKa = 4.92VDD204 pKa = 3.22NLMFAQQGKK213 pKa = 9.63QSGTYY218 pKa = 9.3YY219 pKa = 11.03LDD221 pKa = 4.37ANSHH225 pKa = 5.33TLTMTDD231 pKa = 3.12AQLLHH236 pKa = 6.36NAEE239 pKa = 4.42NDD241 pKa = 3.64DD242 pKa = 4.87CVSNWSDD249 pKa = 3.07MKK251 pKa = 10.79IFSLTEE257 pKa = 3.49DD258 pKa = 3.49TMQLGVLRR266 pKa = 11.84KK267 pKa = 9.7SSCDD271 pKa = 3.01GAAYY275 pKa = 10.33LVFNFISKK283 pKa = 9.91EE284 pKa = 3.95YY285 pKa = 9.64SDD287 pKa = 3.34NWVPEE292 pKa = 4.16EE293 pKa = 4.23TEE295 pKa = 3.87PTIDD299 pKa = 3.71EE300 pKa = 4.68GFDD303 pKa = 3.43PSLEE307 pKa = 4.14AGQLLEE313 pKa = 4.62MLTGGQGSGRR323 pKa = 11.84VWAMDD328 pKa = 3.59AAGNPVDD335 pKa = 4.21WVTAGAGWTVDD346 pKa = 3.43ASSSADD352 pKa = 3.34WGWNSSWSTVASDD365 pKa = 2.58SWIRR369 pKa = 11.84FDD371 pKa = 4.27QWNGMQTYY379 pKa = 8.35TRR381 pKa = 11.84NQSGVEE387 pKa = 3.88TSGTFTINEE396 pKa = 4.09ATNEE400 pKa = 3.88ITLSGEE406 pKa = 4.17VLIQNPGNWMTPTTNTITVIKK427 pKa = 10.62AFDD430 pKa = 4.1DD431 pKa = 4.17YY432 pKa = 9.77QTKK435 pKa = 10.68GIWFGTSYY443 pKa = 11.25DD444 pKa = 3.84SEE446 pKa = 4.63KK447 pKa = 11.28DD448 pKa = 2.84EE449 pKa = 4.38WLAFHH454 pKa = 6.7YY455 pKa = 10.07IIPP458 pKa = 4.18
Molecular weight: 50.49 kDa
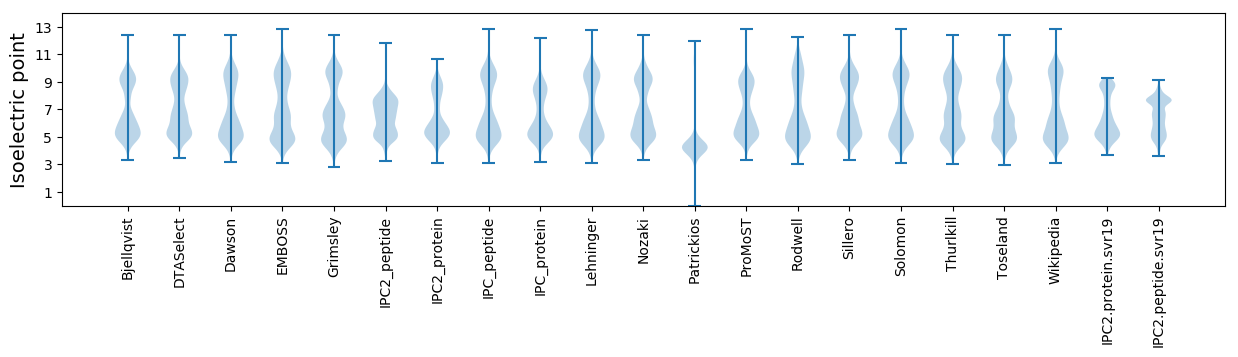
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K2P0B3|K2P0B3_9FLAO Elongation factor G OS=Galbibacter marinus OX=555500 GN=fusA PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVSGFDD22 pKa = 4.27AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.36KK31 pKa = 10.55SLLAPIKK38 pKa = 9.77KK39 pKa = 8.55TGGRR43 pKa = 11.84NSQGKK48 pKa = 6.49MTMRR52 pKa = 11.84YY53 pKa = 9.67LGGGHH58 pKa = 5.72KK59 pKa = 9.83RR60 pKa = 11.84KK61 pKa = 9.98YY62 pKa = 10.35RR63 pKa = 11.84IIDD66 pKa = 3.68FKK68 pKa = 10.73RR69 pKa = 11.84AKK71 pKa = 9.36TGVEE75 pKa = 3.86ATVEE79 pKa = 4.31SIQYY83 pKa = 10.1DD84 pKa = 3.73PNRR87 pKa = 11.84TAFIALIVYY96 pKa = 9.1TDD98 pKa = 3.79GEE100 pKa = 4.25KK101 pKa = 10.59SYY103 pKa = 10.83IIAPNGLQVGQKK115 pKa = 9.39VRR117 pKa = 11.84SGAEE121 pKa = 3.63ATPEE125 pKa = 3.86IGNAMPLSQVPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGAFAQLMARR167 pKa = 11.84EE168 pKa = 4.45GKK170 pKa = 10.09YY171 pKa = 9.05ATIKK175 pKa = 10.52LPSGEE180 pKa = 4.07TRR182 pKa = 11.84YY183 pKa = 10.7VLADD187 pKa = 3.65CFATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.87AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.11PRR245 pKa = 11.84SRR247 pKa = 11.84NGVPAKK253 pKa = 10.34GYY255 pKa = 7.34RR256 pKa = 11.84TRR258 pKa = 11.84AKK260 pKa = 10.32AKK262 pKa = 10.15ASNKK266 pKa = 10.05YY267 pKa = 9.32IVEE270 pKa = 4.08RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVSGFDD22 pKa = 4.27AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.36KK31 pKa = 10.55SLLAPIKK38 pKa = 9.77KK39 pKa = 8.55TGGRR43 pKa = 11.84NSQGKK48 pKa = 6.49MTMRR52 pKa = 11.84YY53 pKa = 9.67LGGGHH58 pKa = 5.72KK59 pKa = 9.83RR60 pKa = 11.84KK61 pKa = 9.98YY62 pKa = 10.35RR63 pKa = 11.84IIDD66 pKa = 3.68FKK68 pKa = 10.73RR69 pKa = 11.84AKK71 pKa = 9.36TGVEE75 pKa = 3.86ATVEE79 pKa = 4.31SIQYY83 pKa = 10.1DD84 pKa = 3.73PNRR87 pKa = 11.84TAFIALIVYY96 pKa = 9.1TDD98 pKa = 3.79GEE100 pKa = 4.25KK101 pKa = 10.59SYY103 pKa = 10.83IIAPNGLQVGQKK115 pKa = 9.39VRR117 pKa = 11.84SGAEE121 pKa = 3.63ATPEE125 pKa = 3.86IGNAMPLSQVPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGAFAQLMARR167 pKa = 11.84EE168 pKa = 4.45GKK170 pKa = 10.09YY171 pKa = 9.05ATIKK175 pKa = 10.52LPSGEE180 pKa = 4.07TRR182 pKa = 11.84YY183 pKa = 10.7VLADD187 pKa = 3.65CFATIGAVSNSDD199 pKa = 3.2HH200 pKa = 6.21QLLVSGKK207 pKa = 9.87AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.11PRR245 pKa = 11.84SRR247 pKa = 11.84NGVPAKK253 pKa = 10.34GYY255 pKa = 7.34RR256 pKa = 11.84TRR258 pKa = 11.84AKK260 pKa = 10.32AKK262 pKa = 10.15ASNKK266 pKa = 10.05YY267 pKa = 9.32IVEE270 pKa = 4.08RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
Molecular weight: 29.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1050357 |
35 |
4820 |
338.9 |
38.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.356 ± 0.043 | 0.73 ± 0.013 |
5.709 ± 0.035 | 6.505 ± 0.038 |
4.892 ± 0.031 | 6.586 ± 0.044 |
1.942 ± 0.023 | 7.676 ± 0.044 |
7.249 ± 0.052 | 9.403 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.27 ± 0.022 | 5.687 ± 0.039 |
3.536 ± 0.025 | 3.976 ± 0.029 |
3.689 ± 0.023 | 6.668 ± 0.033 |
5.574 ± 0.049 | 6.283 ± 0.034 |
1.112 ± 0.016 | 4.157 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
