
Clostridium sp. CAG:217
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
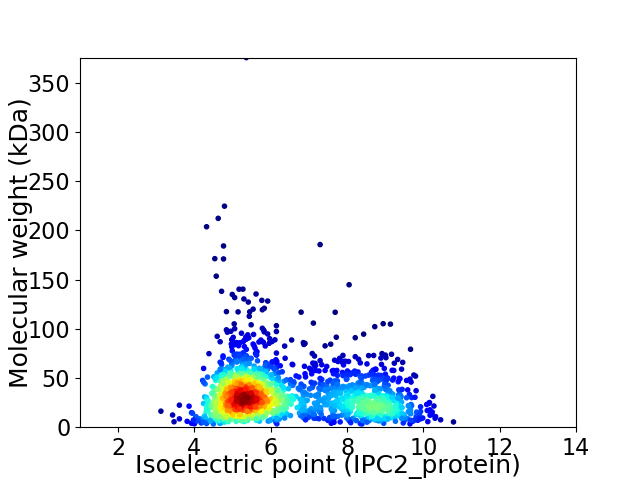
Virtual 2D-PAGE plot for 1746 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6IYX9|R6IYX9_9CLOT Pyruvate kinase OS=Clostridium sp. CAG:217 OX=1262779 GN=BN539_01545 PE=3 SV=1
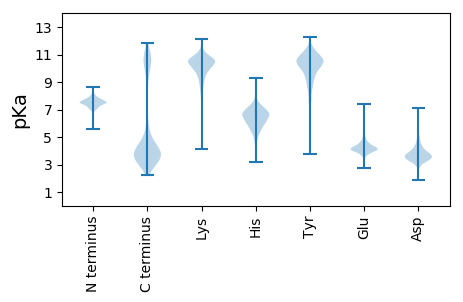
MM1 pKa = 7.38KK2 pKa = 10.25KK3 pKa = 9.67IAIVYY8 pKa = 8.91WSGTGNTAQMAEE20 pKa = 4.17AVAEE24 pKa = 4.33GAGAAGAQANLFTPSDD40 pKa = 4.09FSPAQVKK47 pKa = 10.51DD48 pKa = 3.35YY49 pKa = 11.36DD50 pKa = 4.86AIAFGCPAMGAEE62 pKa = 3.95QLEE65 pKa = 4.49DD66 pKa = 4.38FEE68 pKa = 5.28FEE70 pKa = 4.58PLFLQCAPEE79 pKa = 4.38LNGKK83 pKa = 9.75DD84 pKa = 2.78IALFGSYY91 pKa = 9.01GWGDD95 pKa = 4.25GEE97 pKa = 5.46WMQTWEE103 pKa = 4.65DD104 pKa = 3.42QCKK107 pKa = 10.59DD108 pKa = 3.35LGAHH112 pKa = 6.2LVCNSVIANEE122 pKa = 4.57APDD125 pKa = 4.0DD126 pKa = 3.88TALAACRR133 pKa = 11.84NLGAALAA140 pKa = 4.43
MM1 pKa = 7.38KK2 pKa = 10.25KK3 pKa = 9.67IAIVYY8 pKa = 8.91WSGTGNTAQMAEE20 pKa = 4.17AVAEE24 pKa = 4.33GAGAAGAQANLFTPSDD40 pKa = 4.09FSPAQVKK47 pKa = 10.51DD48 pKa = 3.35YY49 pKa = 11.36DD50 pKa = 4.86AIAFGCPAMGAEE62 pKa = 3.95QLEE65 pKa = 4.49DD66 pKa = 4.38FEE68 pKa = 5.28FEE70 pKa = 4.58PLFLQCAPEE79 pKa = 4.38LNGKK83 pKa = 9.75DD84 pKa = 2.78IALFGSYY91 pKa = 9.01GWGDD95 pKa = 4.25GEE97 pKa = 5.46WMQTWEE103 pKa = 4.65DD104 pKa = 3.42QCKK107 pKa = 10.59DD108 pKa = 3.35LGAHH112 pKa = 6.2LVCNSVIANEE122 pKa = 4.57APDD125 pKa = 4.0DD126 pKa = 3.88TALAACRR133 pKa = 11.84NLGAALAA140 pKa = 4.43
Molecular weight: 14.76 kDa
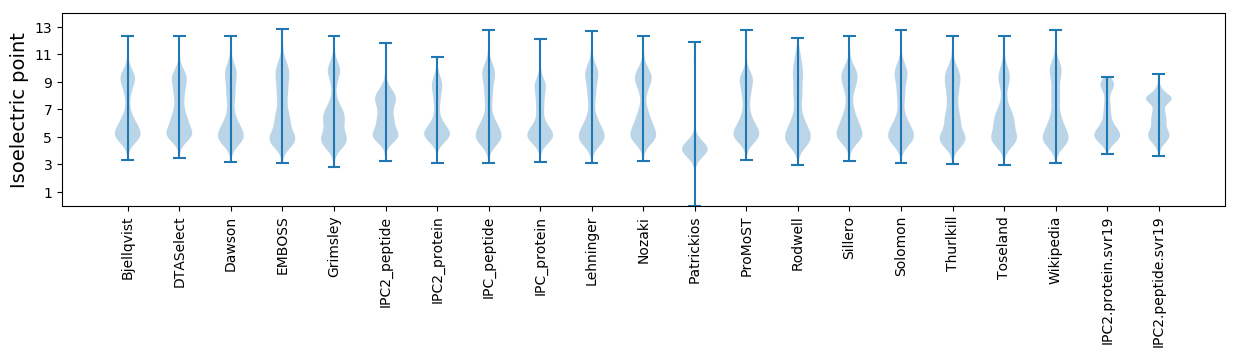
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6J2B6|R6J2B6_9CLOT PS_pyruv_trans domain-containing protein OS=Clostridium sp. CAG:217 OX=1262779 GN=BN539_00136 PE=4 SV=1
MM1 pKa = 7.46AVTHH5 pKa = 6.43GSLRR9 pKa = 11.84HH10 pKa = 5.36TGFNNIHH17 pKa = 5.7QLEE20 pKa = 4.4CSLNGLFLPGFYY32 pKa = 10.42NISGNAAAEE41 pKa = 4.12FFLPIAVEE49 pKa = 4.29NIRR52 pKa = 11.84QRR54 pKa = 11.84SLVISIDD61 pKa = 4.12HH62 pKa = 6.48ICGAAAGLTHH72 pKa = 6.05THH74 pKa = 4.92IQRR77 pKa = 11.84RR78 pKa = 11.84VLMEE82 pKa = 3.96GEE84 pKa = 4.22TPLRR88 pKa = 11.84RR89 pKa = 11.84IQLKK93 pKa = 10.22RR94 pKa = 11.84ADD96 pKa = 3.91SQVQQHH102 pKa = 6.45TIYY105 pKa = 10.8AAQTPVDD112 pKa = 3.43QYY114 pKa = 11.3VFNIRR119 pKa = 11.84KK120 pKa = 8.01IVVYY124 pKa = 10.11NGHH127 pKa = 6.54IRR129 pKa = 11.84QIVQPSVQHH138 pKa = 5.95RR139 pKa = 11.84NGVRR143 pKa = 11.84VLIHH147 pKa = 6.01CQQVAAAFCNSARR160 pKa = 11.84VTAAPCRR167 pKa = 11.84TVYY170 pKa = 10.63QPFARR175 pKa = 11.84GRR177 pKa = 11.84LHH179 pKa = 6.76GCHH182 pKa = 6.56RR183 pKa = 11.84LCKK186 pKa = 9.97QHH188 pKa = 5.9TLVHH192 pKa = 6.34KK193 pKa = 9.37FHH195 pKa = 7.28LIHH198 pKa = 6.87LPSALPXXXLLFRR211 pKa = 11.84SRR213 pKa = 11.84LRR215 pKa = 11.84FFTGIHH221 pKa = 5.84QIQEE225 pKa = 3.75VGIRR229 pKa = 11.84PQAVEE234 pKa = 4.32HH235 pKa = 6.1GVEE238 pKa = 4.23ALIGPNLNGALVALHH253 pKa = 5.03NHH255 pKa = 5.03GRR257 pKa = 11.84AVFHH261 pKa = 6.35TGIVFQPGIQRR272 pKa = 11.84KK273 pKa = 8.24AALCVQRR280 pKa = 11.84TLLCVGEE287 pKa = 4.25EE288 pKa = 3.89NFVFLIHH295 pKa = 6.26FVSSTVVQRR304 pKa = 11.84LHH306 pKa = 5.92TVHH309 pKa = 7.28KK310 pKa = 10.0LILGIQFDD318 pKa = 4.11TGAAPQVHH326 pKa = 6.13TNHH329 pKa = 6.47KK330 pKa = 9.61KK331 pKa = 10.1FRR333 pKa = 11.84SQAFPPLGRR342 pKa = 11.84NRR344 pKa = 11.84KK345 pKa = 9.42APFAVQLLVHH355 pKa = 6.69ICTHH359 pKa = 7.1KK360 pKa = 10.35DD361 pKa = 3.23TQIHH365 pKa = 5.89KK366 pKa = 10.45RR367 pKa = 11.84RR368 pKa = 11.84LLSAKK373 pKa = 9.53INRR376 pKa = 11.84MHH378 pKa = 6.97LWEE381 pKa = 5.45IPPKK385 pKa = 8.97SQVWLGTVMKK395 pKa = 10.72NDD397 pKa = 4.94GIQWNSHH404 pKa = 4.86HH405 pKa = 7.29SYY407 pKa = 11.36LLL409 pKa = 3.78
MM1 pKa = 7.46AVTHH5 pKa = 6.43GSLRR9 pKa = 11.84HH10 pKa = 5.36TGFNNIHH17 pKa = 5.7QLEE20 pKa = 4.4CSLNGLFLPGFYY32 pKa = 10.42NISGNAAAEE41 pKa = 4.12FFLPIAVEE49 pKa = 4.29NIRR52 pKa = 11.84QRR54 pKa = 11.84SLVISIDD61 pKa = 4.12HH62 pKa = 6.48ICGAAAGLTHH72 pKa = 6.05THH74 pKa = 4.92IQRR77 pKa = 11.84RR78 pKa = 11.84VLMEE82 pKa = 3.96GEE84 pKa = 4.22TPLRR88 pKa = 11.84RR89 pKa = 11.84IQLKK93 pKa = 10.22RR94 pKa = 11.84ADD96 pKa = 3.91SQVQQHH102 pKa = 6.45TIYY105 pKa = 10.8AAQTPVDD112 pKa = 3.43QYY114 pKa = 11.3VFNIRR119 pKa = 11.84KK120 pKa = 8.01IVVYY124 pKa = 10.11NGHH127 pKa = 6.54IRR129 pKa = 11.84QIVQPSVQHH138 pKa = 5.95RR139 pKa = 11.84NGVRR143 pKa = 11.84VLIHH147 pKa = 6.01CQQVAAAFCNSARR160 pKa = 11.84VTAAPCRR167 pKa = 11.84TVYY170 pKa = 10.63QPFARR175 pKa = 11.84GRR177 pKa = 11.84LHH179 pKa = 6.76GCHH182 pKa = 6.56RR183 pKa = 11.84LCKK186 pKa = 9.97QHH188 pKa = 5.9TLVHH192 pKa = 6.34KK193 pKa = 9.37FHH195 pKa = 7.28LIHH198 pKa = 6.87LPSALPXXXLLFRR211 pKa = 11.84SRR213 pKa = 11.84LRR215 pKa = 11.84FFTGIHH221 pKa = 5.84QIQEE225 pKa = 3.75VGIRR229 pKa = 11.84PQAVEE234 pKa = 4.32HH235 pKa = 6.1GVEE238 pKa = 4.23ALIGPNLNGALVALHH253 pKa = 5.03NHH255 pKa = 5.03GRR257 pKa = 11.84AVFHH261 pKa = 6.35TGIVFQPGIQRR272 pKa = 11.84KK273 pKa = 8.24AALCVQRR280 pKa = 11.84TLLCVGEE287 pKa = 4.25EE288 pKa = 3.89NFVFLIHH295 pKa = 6.26FVSSTVVQRR304 pKa = 11.84LHH306 pKa = 5.92TVHH309 pKa = 7.28KK310 pKa = 10.0LILGIQFDD318 pKa = 4.11TGAAPQVHH326 pKa = 6.13TNHH329 pKa = 6.47KK330 pKa = 9.61KK331 pKa = 10.1FRR333 pKa = 11.84SQAFPPLGRR342 pKa = 11.84NRR344 pKa = 11.84KK345 pKa = 9.42APFAVQLLVHH355 pKa = 6.69ICTHH359 pKa = 7.1KK360 pKa = 10.35DD361 pKa = 3.23TQIHH365 pKa = 5.89KK366 pKa = 10.45RR367 pKa = 11.84RR368 pKa = 11.84LLSAKK373 pKa = 9.53INRR376 pKa = 11.84MHH378 pKa = 6.97LWEE381 pKa = 5.45IPPKK385 pKa = 8.97SQVWLGTVMKK395 pKa = 10.72NDD397 pKa = 4.94GIQWNSHH404 pKa = 4.86HH405 pKa = 7.29SYY407 pKa = 11.36LLL409 pKa = 3.78
Molecular weight: 45.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
554469 |
29 |
3359 |
317.6 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.52 ± 0.071 | 1.778 ± 0.027 |
6.02 ± 0.05 | 5.711 ± 0.063 |
3.973 ± 0.043 | 7.523 ± 0.054 |
1.923 ± 0.031 | 5.776 ± 0.065 |
6.003 ± 0.052 | 9.195 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.67 ± 0.027 | 3.909 ± 0.043 |
3.844 ± 0.041 | 3.645 ± 0.041 |
4.872 ± 0.054 | 5.193 ± 0.051 |
6.119 ± 0.065 | 7.473 ± 0.051 |
0.872 ± 0.023 | 3.966 ± 0.047 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
