
Rhizobium oryzae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
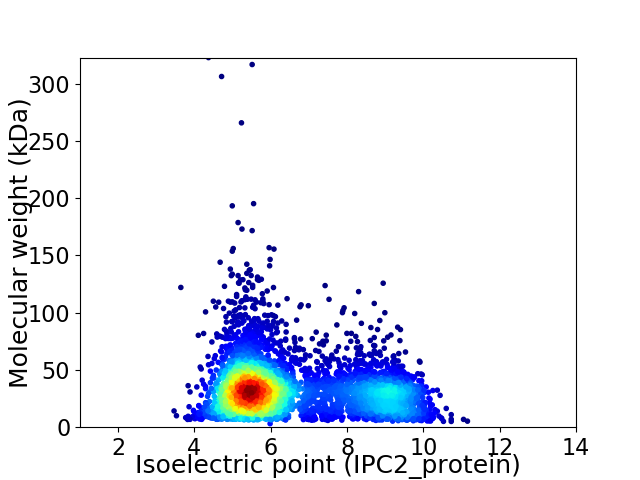
Virtual 2D-PAGE plot for 4627 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q9AUC5|A0A1Q9AUC5_9RHIZ Protein SlyX homolog OS=Rhizobium oryzae OX=464029 GN=slyX PE=3 SV=1
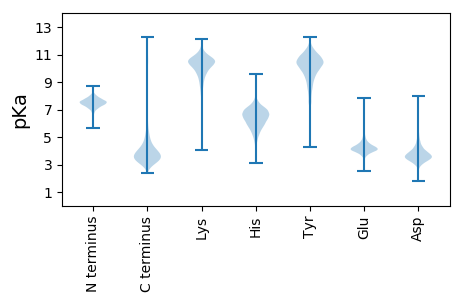
MM1 pKa = 6.7STEE4 pKa = 4.24TKK6 pKa = 9.57VLPMISGTSANDD18 pKa = 3.68YY19 pKa = 10.23IASTGSGATLVGGAGNDD36 pKa = 3.59LYY38 pKa = 11.07FVTSADD44 pKa = 3.47DD45 pKa = 4.19VIIEE49 pKa = 4.43KK50 pKa = 10.06PGEE53 pKa = 4.56GIDD56 pKa = 4.06SVITYY61 pKa = 7.49RR62 pKa = 11.84TSYY65 pKa = 9.9TLGDD69 pKa = 3.5NVEE72 pKa = 4.21NLSVLGNDD80 pKa = 4.11LYY82 pKa = 11.64GVGNALDD89 pKa = 3.75NVLVGGSGRR98 pKa = 11.84QTLVGGAGNDD108 pKa = 3.7TLTGGAGSDD117 pKa = 3.51IFVLTQGGGSDD128 pKa = 4.0TITDD132 pKa = 4.38FDD134 pKa = 4.8ASDD137 pKa = 3.28IVQLGVYY144 pKa = 9.18GFKK147 pKa = 10.93NFAEE151 pKa = 4.48VSDD154 pKa = 5.48SLVQDD159 pKa = 3.56NANVVLKK166 pKa = 11.06LNDD169 pKa = 3.53GSQLTFLNKK178 pKa = 9.77TIADD182 pKa = 3.97FQSSNFQYY190 pKa = 11.55ALDD193 pKa = 4.03TSHH196 pKa = 7.34LKK198 pKa = 8.15QTFADD203 pKa = 3.91EE204 pKa = 4.8FNTLSLQQEE213 pKa = 4.28GGMWRR218 pKa = 11.84TWHH221 pKa = 6.55NNGLSDD227 pKa = 5.24GGQAYY232 pKa = 10.42LNPTNSTGVNPFSVTDD248 pKa = 4.08GVLTIHH254 pKa = 7.49ADD256 pKa = 3.66RR257 pKa = 11.84ASAEE261 pKa = 4.19TAALTGKK268 pKa = 9.73EE269 pKa = 4.05FTSGIMSTRR278 pKa = 11.84DD279 pKa = 3.31TFSQTYY285 pKa = 9.57GYY287 pKa = 10.79FEE289 pKa = 4.27IRR291 pKa = 11.84ADD293 pKa = 3.74LPSQKK298 pKa = 10.12GVCPAFWLLPANGSWPPEE316 pKa = 3.67LDD318 pKa = 3.2VFEE321 pKa = 4.69QVGNDD326 pKa = 3.45PSKK329 pKa = 11.33VYY331 pKa = 9.19LTAHH335 pKa = 6.16SNSTGTPTATGISSWIGDD353 pKa = 3.91TGEE356 pKa = 4.18GMHH359 pKa = 6.67TYY361 pKa = 10.87GLLWTKK367 pKa = 10.89DD368 pKa = 3.38VLVWYY373 pKa = 10.17LDD375 pKa = 3.26GVEE378 pKa = 4.72AYY380 pKa = 10.35RR381 pKa = 11.84IATPADD387 pKa = 3.33MHH389 pKa = 5.61QAMYY393 pKa = 10.7LVTNMSVGGGFAGSTDD409 pKa = 3.33ASFTGADD416 pKa = 3.54FKK418 pKa = 10.92IDD420 pKa = 3.92YY421 pKa = 9.74IHH423 pKa = 7.88AYY425 pKa = 8.44QLEE428 pKa = 4.61EE429 pKa = 3.96NLGTRR434 pKa = 11.84QSITGEE440 pKa = 3.79KK441 pKa = 9.72WSITLDD447 pKa = 3.54NNGDD451 pKa = 3.93DD452 pKa = 3.81VTLTGTLNINATGNAKK468 pKa = 10.63DD469 pKa = 4.31NILIGNSGNNTLDD482 pKa = 3.54GKK484 pKa = 11.01VGADD488 pKa = 3.46TMIGGQGDD496 pKa = 3.55DD497 pKa = 3.72TYY499 pKa = 11.34IVDD502 pKa = 3.53NAGDD506 pKa = 3.83VVVEE510 pKa = 4.15NANEE514 pKa = 4.1GTDD517 pKa = 3.46TVRR520 pKa = 11.84ASISYY525 pKa = 9.59VLGANVEE532 pKa = 4.32KK533 pKa = 11.02LFLTGTDD540 pKa = 3.48NLHH543 pKa = 5.44GTGNEE548 pKa = 4.15LDD550 pKa = 3.4NTIYY554 pKa = 10.96GNSGDD559 pKa = 3.96NVINGAGGADD569 pKa = 3.72VMIGGAGNDD578 pKa = 2.99IYY580 pKa = 11.65YY581 pKa = 10.36VDD583 pKa = 3.98NPGDD587 pKa = 4.09IVTEE591 pKa = 4.41WSNGGIDD598 pKa = 3.62TVYY601 pKa = 11.21SSISYY606 pKa = 10.08ALPNQVEE613 pKa = 4.31NLTLTGSDD621 pKa = 4.14NIDD624 pKa = 3.26ATGNWMDD631 pKa = 5.24NILIGNDD638 pKa = 3.13GNNVLRR644 pKa = 11.84GMGGTDD650 pKa = 3.19TLIGGKK656 pKa = 10.23GDD658 pKa = 3.21DD659 pKa = 3.73TYY661 pKa = 11.93YY662 pKa = 11.03VDD664 pKa = 5.13SDD666 pKa = 3.54DD667 pKa = 4.83TIIEE671 pKa = 4.26KK672 pKa = 10.64ADD674 pKa = 3.35GGIDD678 pKa = 3.35TVYY681 pKa = 11.04ASSSYY686 pKa = 9.6TLSANVEE693 pKa = 4.28TLILTGNWTSNGTGNNLDD711 pKa = 3.76NLLIGNSCANTLDD724 pKa = 3.71GGRR727 pKa = 11.84GADD730 pKa = 3.58TMIGGAGNDD739 pKa = 3.55TYY741 pKa = 11.24FVDD744 pKa = 3.76NPGDD748 pKa = 3.65IVIEE752 pKa = 4.1NAGEE756 pKa = 4.23GVDD759 pKa = 3.65RR760 pKa = 11.84VYY762 pKa = 11.57SSISYY767 pKa = 8.6TLTANVEE774 pKa = 4.16MLLLTGKK781 pKa = 10.58ADD783 pKa = 4.28LNATGNSLSNTIYY796 pKa = 11.13GNDD799 pKa = 3.63GNNIIDD805 pKa = 4.33GGTGADD811 pKa = 3.34TMSGQGGNDD820 pKa = 3.06TYY822 pKa = 11.61YY823 pKa = 11.16VDD825 pKa = 4.55NIYY828 pKa = 11.17DD829 pKa = 5.15KK830 pKa = 10.83IIEE833 pKa = 4.14WSNGGIDD840 pKa = 3.65TVLSSVSYY848 pKa = 10.77SLADD852 pKa = 3.37NVEE855 pKa = 3.82NLTLIGLANLNATGNWQDD873 pKa = 4.11NILIGNAGNNILDD886 pKa = 3.89GSKK889 pKa = 10.84GADD892 pKa = 3.36LMKK895 pKa = 10.82GGAGDD900 pKa = 3.36DD901 pKa = 3.8TYY903 pKa = 11.24IVEE906 pKa = 4.32NVGDD910 pKa = 3.91VVVEE914 pKa = 4.1NPGEE918 pKa = 4.44GIDD921 pKa = 4.12TVKK924 pKa = 10.64SWISYY929 pKa = 9.81RR930 pKa = 11.84LTDD933 pKa = 3.74NVEE936 pKa = 3.96KK937 pKa = 11.06LMLQSAGNINGYY949 pKa = 10.88GNDD952 pKa = 4.0LDD954 pKa = 3.95NTIVGSIGNNIIDD967 pKa = 4.26GGAGADD973 pKa = 3.5RR974 pKa = 11.84MQGGLGNDD982 pKa = 3.44TYY984 pKa = 11.78YY985 pKa = 11.23VDD987 pKa = 3.82NPGDD991 pKa = 4.6LVIEE995 pKa = 4.47WSGEE999 pKa = 4.13GNDD1002 pKa = 3.9TVYY1005 pKa = 11.34SSISYY1010 pKa = 8.48TLVNNVEE1017 pKa = 3.96NLILTGTANLDD1028 pKa = 3.48GCGNGLDD1035 pKa = 3.68NTLIGNDD1042 pKa = 3.39GNNRR1046 pKa = 11.84LDD1048 pKa = 3.71GGAGNDD1054 pKa = 3.35ILQGGLGVDD1063 pKa = 3.83ILNGGAGNDD1072 pKa = 3.3TFVFKK1077 pKa = 9.84TAADD1081 pKa = 4.03SAVGTPDD1088 pKa = 4.62TINDD1092 pKa = 3.73FQKK1095 pKa = 11.34GDD1097 pKa = 3.86IIDD1100 pKa = 4.16LRR1102 pKa = 11.84DD1103 pKa = 3.54MYY1105 pKa = 11.06AGTMTFLGTGDD1116 pKa = 3.68FTGHH1120 pKa = 5.97SGEE1123 pKa = 4.35VQISAYY1129 pKa = 10.55GKK1131 pKa = 8.77GTMINIDD1138 pKa = 3.81LDD1140 pKa = 3.9GDD1142 pKa = 3.72KK1143 pKa = 10.99HH1144 pKa = 7.55IDD1146 pKa = 3.45SQIFVANLQPSNFASQGGIFLLL1168 pKa = 4.47
MM1 pKa = 6.7STEE4 pKa = 4.24TKK6 pKa = 9.57VLPMISGTSANDD18 pKa = 3.68YY19 pKa = 10.23IASTGSGATLVGGAGNDD36 pKa = 3.59LYY38 pKa = 11.07FVTSADD44 pKa = 3.47DD45 pKa = 4.19VIIEE49 pKa = 4.43KK50 pKa = 10.06PGEE53 pKa = 4.56GIDD56 pKa = 4.06SVITYY61 pKa = 7.49RR62 pKa = 11.84TSYY65 pKa = 9.9TLGDD69 pKa = 3.5NVEE72 pKa = 4.21NLSVLGNDD80 pKa = 4.11LYY82 pKa = 11.64GVGNALDD89 pKa = 3.75NVLVGGSGRR98 pKa = 11.84QTLVGGAGNDD108 pKa = 3.7TLTGGAGSDD117 pKa = 3.51IFVLTQGGGSDD128 pKa = 4.0TITDD132 pKa = 4.38FDD134 pKa = 4.8ASDD137 pKa = 3.28IVQLGVYY144 pKa = 9.18GFKK147 pKa = 10.93NFAEE151 pKa = 4.48VSDD154 pKa = 5.48SLVQDD159 pKa = 3.56NANVVLKK166 pKa = 11.06LNDD169 pKa = 3.53GSQLTFLNKK178 pKa = 9.77TIADD182 pKa = 3.97FQSSNFQYY190 pKa = 11.55ALDD193 pKa = 4.03TSHH196 pKa = 7.34LKK198 pKa = 8.15QTFADD203 pKa = 3.91EE204 pKa = 4.8FNTLSLQQEE213 pKa = 4.28GGMWRR218 pKa = 11.84TWHH221 pKa = 6.55NNGLSDD227 pKa = 5.24GGQAYY232 pKa = 10.42LNPTNSTGVNPFSVTDD248 pKa = 4.08GVLTIHH254 pKa = 7.49ADD256 pKa = 3.66RR257 pKa = 11.84ASAEE261 pKa = 4.19TAALTGKK268 pKa = 9.73EE269 pKa = 4.05FTSGIMSTRR278 pKa = 11.84DD279 pKa = 3.31TFSQTYY285 pKa = 9.57GYY287 pKa = 10.79FEE289 pKa = 4.27IRR291 pKa = 11.84ADD293 pKa = 3.74LPSQKK298 pKa = 10.12GVCPAFWLLPANGSWPPEE316 pKa = 3.67LDD318 pKa = 3.2VFEE321 pKa = 4.69QVGNDD326 pKa = 3.45PSKK329 pKa = 11.33VYY331 pKa = 9.19LTAHH335 pKa = 6.16SNSTGTPTATGISSWIGDD353 pKa = 3.91TGEE356 pKa = 4.18GMHH359 pKa = 6.67TYY361 pKa = 10.87GLLWTKK367 pKa = 10.89DD368 pKa = 3.38VLVWYY373 pKa = 10.17LDD375 pKa = 3.26GVEE378 pKa = 4.72AYY380 pKa = 10.35RR381 pKa = 11.84IATPADD387 pKa = 3.33MHH389 pKa = 5.61QAMYY393 pKa = 10.7LVTNMSVGGGFAGSTDD409 pKa = 3.33ASFTGADD416 pKa = 3.54FKK418 pKa = 10.92IDD420 pKa = 3.92YY421 pKa = 9.74IHH423 pKa = 7.88AYY425 pKa = 8.44QLEE428 pKa = 4.61EE429 pKa = 3.96NLGTRR434 pKa = 11.84QSITGEE440 pKa = 3.79KK441 pKa = 9.72WSITLDD447 pKa = 3.54NNGDD451 pKa = 3.93DD452 pKa = 3.81VTLTGTLNINATGNAKK468 pKa = 10.63DD469 pKa = 4.31NILIGNSGNNTLDD482 pKa = 3.54GKK484 pKa = 11.01VGADD488 pKa = 3.46TMIGGQGDD496 pKa = 3.55DD497 pKa = 3.72TYY499 pKa = 11.34IVDD502 pKa = 3.53NAGDD506 pKa = 3.83VVVEE510 pKa = 4.15NANEE514 pKa = 4.1GTDD517 pKa = 3.46TVRR520 pKa = 11.84ASISYY525 pKa = 9.59VLGANVEE532 pKa = 4.32KK533 pKa = 11.02LFLTGTDD540 pKa = 3.48NLHH543 pKa = 5.44GTGNEE548 pKa = 4.15LDD550 pKa = 3.4NTIYY554 pKa = 10.96GNSGDD559 pKa = 3.96NVINGAGGADD569 pKa = 3.72VMIGGAGNDD578 pKa = 2.99IYY580 pKa = 11.65YY581 pKa = 10.36VDD583 pKa = 3.98NPGDD587 pKa = 4.09IVTEE591 pKa = 4.41WSNGGIDD598 pKa = 3.62TVYY601 pKa = 11.21SSISYY606 pKa = 10.08ALPNQVEE613 pKa = 4.31NLTLTGSDD621 pKa = 4.14NIDD624 pKa = 3.26ATGNWMDD631 pKa = 5.24NILIGNDD638 pKa = 3.13GNNVLRR644 pKa = 11.84GMGGTDD650 pKa = 3.19TLIGGKK656 pKa = 10.23GDD658 pKa = 3.21DD659 pKa = 3.73TYY661 pKa = 11.93YY662 pKa = 11.03VDD664 pKa = 5.13SDD666 pKa = 3.54DD667 pKa = 4.83TIIEE671 pKa = 4.26KK672 pKa = 10.64ADD674 pKa = 3.35GGIDD678 pKa = 3.35TVYY681 pKa = 11.04ASSSYY686 pKa = 9.6TLSANVEE693 pKa = 4.28TLILTGNWTSNGTGNNLDD711 pKa = 3.76NLLIGNSCANTLDD724 pKa = 3.71GGRR727 pKa = 11.84GADD730 pKa = 3.58TMIGGAGNDD739 pKa = 3.55TYY741 pKa = 11.24FVDD744 pKa = 3.76NPGDD748 pKa = 3.65IVIEE752 pKa = 4.1NAGEE756 pKa = 4.23GVDD759 pKa = 3.65RR760 pKa = 11.84VYY762 pKa = 11.57SSISYY767 pKa = 8.6TLTANVEE774 pKa = 4.16MLLLTGKK781 pKa = 10.58ADD783 pKa = 4.28LNATGNSLSNTIYY796 pKa = 11.13GNDD799 pKa = 3.63GNNIIDD805 pKa = 4.33GGTGADD811 pKa = 3.34TMSGQGGNDD820 pKa = 3.06TYY822 pKa = 11.61YY823 pKa = 11.16VDD825 pKa = 4.55NIYY828 pKa = 11.17DD829 pKa = 5.15KK830 pKa = 10.83IIEE833 pKa = 4.14WSNGGIDD840 pKa = 3.65TVLSSVSYY848 pKa = 10.77SLADD852 pKa = 3.37NVEE855 pKa = 3.82NLTLIGLANLNATGNWQDD873 pKa = 4.11NILIGNAGNNILDD886 pKa = 3.89GSKK889 pKa = 10.84GADD892 pKa = 3.36LMKK895 pKa = 10.82GGAGDD900 pKa = 3.36DD901 pKa = 3.8TYY903 pKa = 11.24IVEE906 pKa = 4.32NVGDD910 pKa = 3.91VVVEE914 pKa = 4.1NPGEE918 pKa = 4.44GIDD921 pKa = 4.12TVKK924 pKa = 10.64SWISYY929 pKa = 9.81RR930 pKa = 11.84LTDD933 pKa = 3.74NVEE936 pKa = 3.96KK937 pKa = 11.06LMLQSAGNINGYY949 pKa = 10.88GNDD952 pKa = 4.0LDD954 pKa = 3.95NTIVGSIGNNIIDD967 pKa = 4.26GGAGADD973 pKa = 3.5RR974 pKa = 11.84MQGGLGNDD982 pKa = 3.44TYY984 pKa = 11.78YY985 pKa = 11.23VDD987 pKa = 3.82NPGDD991 pKa = 4.6LVIEE995 pKa = 4.47WSGEE999 pKa = 4.13GNDD1002 pKa = 3.9TVYY1005 pKa = 11.34SSISYY1010 pKa = 8.48TLVNNVEE1017 pKa = 3.96NLILTGTANLDD1028 pKa = 3.48GCGNGLDD1035 pKa = 3.68NTLIGNDD1042 pKa = 3.39GNNRR1046 pKa = 11.84LDD1048 pKa = 3.71GGAGNDD1054 pKa = 3.35ILQGGLGVDD1063 pKa = 3.83ILNGGAGNDD1072 pKa = 3.3TFVFKK1077 pKa = 9.84TAADD1081 pKa = 4.03SAVGTPDD1088 pKa = 4.62TINDD1092 pKa = 3.73FQKK1095 pKa = 11.34GDD1097 pKa = 3.86IIDD1100 pKa = 4.16LRR1102 pKa = 11.84DD1103 pKa = 3.54MYY1105 pKa = 11.06AGTMTFLGTGDD1116 pKa = 3.68FTGHH1120 pKa = 5.97SGEE1123 pKa = 4.35VQISAYY1129 pKa = 10.55GKK1131 pKa = 8.77GTMINIDD1138 pKa = 3.81LDD1140 pKa = 3.9GDD1142 pKa = 3.72KK1143 pKa = 10.99HH1144 pKa = 7.55IDD1146 pKa = 3.45SQIFVANLQPSNFASQGGIFLLL1168 pKa = 4.47
Molecular weight: 122.05 kDa
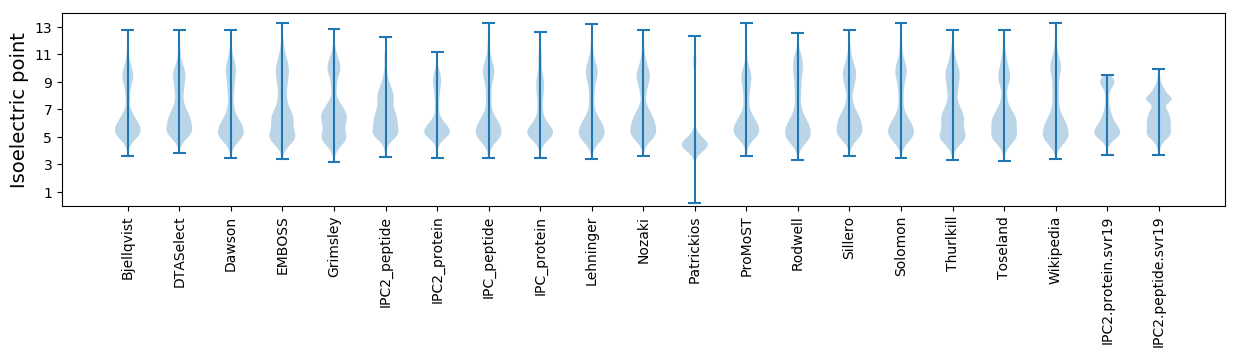
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q9AT48|A0A1Q9AT48_9RHIZ 3-oxoacyl-[acyl-carrier-protein] synthase 2 OS=Rhizobium oryzae OX=464029 GN=BJF93_17060 PE=3 SV=1
MM1 pKa = 7.42FWRR4 pKa = 11.84SAAFAVAWAVGHH16 pKa = 5.95ASKK19 pKa = 10.57RR20 pKa = 11.84RR21 pKa = 11.84PATKK25 pKa = 8.71SAMMTHH31 pKa = 7.01HH32 pKa = 6.61VRR34 pKa = 11.84ILLITRR40 pKa = 11.84PILPLGKK47 pKa = 10.02KK48 pKa = 10.15FKK50 pKa = 10.84LFLKK54 pKa = 10.3APRR57 pKa = 11.84RR58 pKa = 11.84ASS60 pKa = 3.14
MM1 pKa = 7.42FWRR4 pKa = 11.84SAAFAVAWAVGHH16 pKa = 5.95ASKK19 pKa = 10.57RR20 pKa = 11.84RR21 pKa = 11.84PATKK25 pKa = 8.71SAMMTHH31 pKa = 7.01HH32 pKa = 6.61VRR34 pKa = 11.84ILLITRR40 pKa = 11.84PILPLGKK47 pKa = 10.02KK48 pKa = 10.15FKK50 pKa = 10.84LFLKK54 pKa = 10.3APRR57 pKa = 11.84RR58 pKa = 11.84ASS60 pKa = 3.14
Molecular weight: 6.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1480217 |
28 |
3204 |
319.9 |
34.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.825 ± 0.055 | 0.76 ± 0.01 |
5.711 ± 0.032 | 5.639 ± 0.032 |
3.78 ± 0.021 | 8.373 ± 0.031 |
2.039 ± 0.015 | 5.345 ± 0.025 |
3.278 ± 0.029 | 10.503 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.527 ± 0.017 | 2.486 ± 0.023 |
5.069 ± 0.028 | 3.198 ± 0.021 |
7.076 ± 0.036 | 5.564 ± 0.027 |
5.288 ± 0.035 | 7.159 ± 0.026 |
1.229 ± 0.014 | 2.152 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
