
Beihai sobemo-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
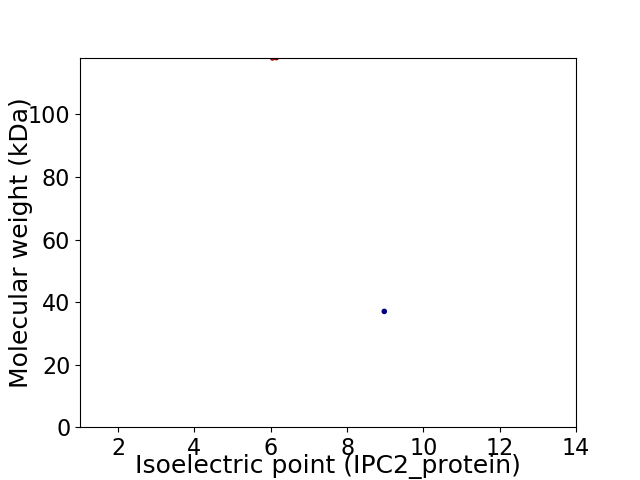
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
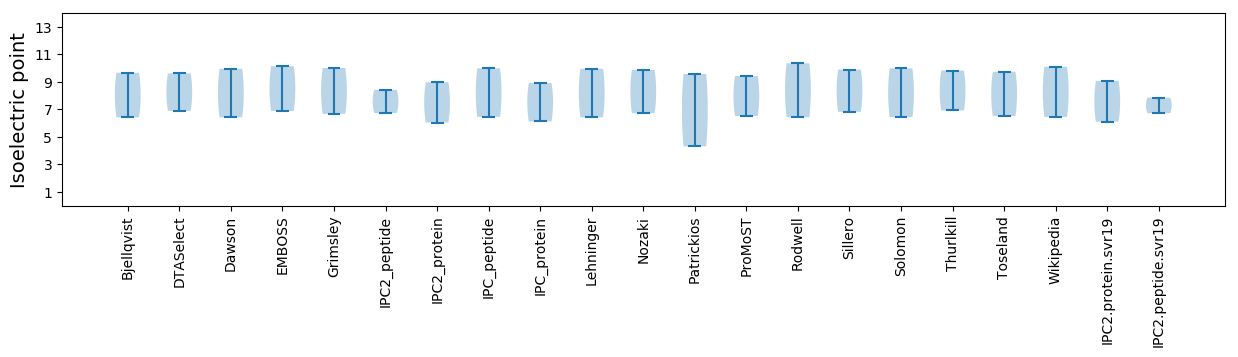
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEA0|A0A1L3KEA0_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 5 OX=1922702 PE=4 SV=1
MM1 pKa = 7.69ALLLSRR7 pKa = 11.84VQGADD12 pKa = 2.94TKK14 pKa = 10.84AVKK17 pKa = 10.29KK18 pKa = 10.57VVTSFGLEE26 pKa = 3.66PGAARR31 pKa = 11.84AYY33 pKa = 10.56LHH35 pKa = 6.84LGLLFGIYY43 pKa = 10.29KK44 pKa = 9.2LVCARR49 pKa = 11.84KK50 pKa = 8.91EE51 pKa = 3.91LCPPIARR58 pKa = 11.84ACLRR62 pKa = 11.84VLYY65 pKa = 9.55YY66 pKa = 9.36VVKK69 pKa = 9.48MSSPGVSLVSWLKK82 pKa = 9.44EE83 pKa = 3.39RR84 pKa = 11.84LRR86 pKa = 11.84RR87 pKa = 11.84GRR89 pKa = 11.84IADD92 pKa = 4.0CVQGGPPTKK101 pKa = 9.9HH102 pKa = 6.8GYY104 pKa = 9.14FRR106 pKa = 11.84WNGKK110 pKa = 9.96DD111 pKa = 3.63FVRR114 pKa = 11.84HH115 pKa = 5.34SDD117 pKa = 3.34EE118 pKa = 4.49SISRR122 pKa = 11.84VKK124 pKa = 9.93MLRR127 pKa = 11.84GTTTIDD133 pKa = 3.02AYY135 pKa = 10.38GQEE138 pKa = 4.02STAYY142 pKa = 8.56NWEE145 pKa = 3.82ILEE148 pKa = 4.06PARR151 pKa = 11.84PKK153 pKa = 10.67DD154 pKa = 3.65LSPGEE159 pKa = 4.19SALEE163 pKa = 4.05GLSSNARR170 pKa = 11.84PSPLEE175 pKa = 4.0KK176 pKa = 10.6GHH178 pKa = 6.18VLVHH182 pKa = 6.97DD183 pKa = 5.18SILNLFSTGLVLNNILIMSRR203 pKa = 11.84HH204 pKa = 4.61KK205 pKa = 9.45TASLTHH211 pKa = 5.78LVVRR215 pKa = 11.84GTNTTAVAGRR225 pKa = 11.84QSGVRR230 pKa = 11.84ISLDD234 pKa = 2.85RR235 pKa = 11.84AYY237 pKa = 11.35YY238 pKa = 10.69LDD240 pKa = 5.66DD241 pKa = 4.41FGAPKK246 pKa = 9.53WDD248 pKa = 4.14EE249 pKa = 4.0KK250 pKa = 11.21RR251 pKa = 11.84HH252 pKa = 5.02TCTFLDD258 pKa = 4.92FMAIPLDD265 pKa = 3.63KK266 pKa = 11.3DD267 pKa = 4.02EE268 pKa = 5.66ISMIGVKK275 pKa = 8.74SLKK278 pKa = 10.83DD279 pKa = 3.21KK280 pKa = 11.08DD281 pKa = 3.11ITRR284 pKa = 11.84NYY286 pKa = 8.76EE287 pKa = 3.93LEE289 pKa = 4.5RR290 pKa = 11.84GTITYY295 pKa = 8.1ATDD298 pKa = 3.42DD299 pKa = 3.73FGNVVKK305 pKa = 10.74EE306 pKa = 4.04EE307 pKa = 4.15GSIPEE312 pKa = 4.36AEE314 pKa = 4.07LHH316 pKa = 5.42IHH318 pKa = 6.3NLGLALAHH326 pKa = 6.73IVSQPGASSSGVGVVQNGYY345 pKa = 10.03KK346 pKa = 10.21LAGMWLGQPAHH357 pKa = 6.15SLKK360 pKa = 10.27KK361 pKa = 10.16HH362 pKa = 5.84RR363 pKa = 11.84GKK365 pKa = 10.97ANLFMHH371 pKa = 7.04TDD373 pKa = 3.69AIMANLEE380 pKa = 3.85QMGLYY385 pKa = 9.37KK386 pKa = 10.56HH387 pKa = 6.28PLIEE391 pKa = 6.31RR392 pKa = 11.84IQQWSDD398 pKa = 2.92SLQTPTSLPEE408 pKa = 3.91APGEE412 pKa = 4.07SRR414 pKa = 11.84EE415 pKa = 4.34SKK417 pKa = 10.43KK418 pKa = 10.46EE419 pKa = 3.32RR420 pKa = 11.84AARR423 pKa = 11.84KK424 pKa = 7.77WFEE427 pKa = 3.87YY428 pKa = 8.17YY429 pKa = 9.75QEE431 pKa = 4.58YY432 pKa = 10.6KK433 pKa = 10.75DD434 pKa = 3.9EE435 pKa = 4.89PYY437 pKa = 11.02EE438 pKa = 4.27AMEE441 pKa = 4.15EE442 pKa = 4.2DD443 pKa = 3.89EE444 pKa = 4.35EE445 pKa = 4.4EE446 pKa = 4.49EE447 pKa = 4.29YY448 pKa = 11.16EE449 pKa = 4.24KK450 pKa = 11.23VYY452 pKa = 10.72GRR454 pKa = 11.84SGHH457 pKa = 6.78ARR459 pKa = 11.84TQPAVALPKK468 pKa = 10.39AVAAPGPRR476 pKa = 11.84IPPRR480 pKa = 11.84ARR482 pKa = 11.84HH483 pKa = 5.84PGEE486 pKa = 4.12SAAEE490 pKa = 3.82RR491 pKa = 11.84ALEE494 pKa = 3.81ITRR497 pKa = 11.84QNMLLDD503 pKa = 3.34IDD505 pKa = 4.98RR506 pKa = 11.84NVKK509 pKa = 8.06SWKK512 pKa = 10.05YY513 pKa = 10.61GILHH517 pKa = 6.41EE518 pKa = 4.31EE519 pKa = 4.06APRR522 pKa = 11.84VLRR525 pKa = 11.84EE526 pKa = 3.25ISYY529 pKa = 10.23YY530 pKa = 10.32RR531 pKa = 11.84EE532 pKa = 3.58EE533 pKa = 3.82ATADD537 pKa = 3.06VRR539 pKa = 11.84EE540 pKa = 4.07RR541 pKa = 11.84LKK543 pKa = 11.14AVLATPSNALLRR555 pKa = 11.84DD556 pKa = 3.65YY557 pKa = 9.51MKK559 pKa = 10.36KK560 pKa = 10.21VSPMWSPGEE569 pKa = 3.92EE570 pKa = 4.2GEE572 pKa = 5.09EE573 pKa = 3.86ILDD576 pKa = 3.94RR577 pKa = 11.84DD578 pKa = 3.65GNPYY582 pKa = 9.25FRR584 pKa = 11.84RR585 pKa = 11.84VGSYY589 pKa = 10.63EE590 pKa = 3.62PTRR593 pKa = 11.84TAKK596 pKa = 9.76AKK598 pKa = 10.15KK599 pKa = 9.36YY600 pKa = 10.2RR601 pKa = 11.84EE602 pKa = 4.22EE603 pKa = 3.89EE604 pKa = 3.56TDD606 pKa = 3.25LQKK609 pKa = 11.0RR610 pKa = 11.84LRR612 pKa = 11.84EE613 pKa = 3.93LANRR617 pKa = 11.84YY618 pKa = 8.81YY619 pKa = 11.07ADD621 pKa = 4.24KK622 pKa = 11.28LHH624 pKa = 6.21GCKK627 pKa = 9.88KK628 pKa = 10.0GEE630 pKa = 3.82YY631 pKa = 9.57RR632 pKa = 11.84IPTSSKK638 pKa = 10.48KK639 pKa = 10.85NIDD642 pKa = 3.6RR643 pKa = 11.84SLKK646 pKa = 9.86AQAALAVAGPPQLTSEE662 pKa = 4.1QEE664 pKa = 4.23EE665 pKa = 4.32EE666 pKa = 4.04FEE668 pKa = 4.02AAVRR672 pKa = 11.84LVRR675 pKa = 11.84EE676 pKa = 4.32KK677 pKa = 10.87YY678 pKa = 9.57KK679 pKa = 11.02AGIGDD684 pKa = 4.19EE685 pKa = 4.45PIKK688 pKa = 10.2TYY690 pKa = 10.92LEE692 pKa = 4.13EE693 pKa = 5.78GDD695 pKa = 3.83YY696 pKa = 11.64GFLKK700 pKa = 10.14TFLGFEE706 pKa = 4.53DD707 pKa = 4.05KK708 pKa = 11.14SSGVSARR715 pKa = 11.84FRR717 pKa = 11.84NMKK720 pKa = 8.88KK721 pKa = 9.9AAWVKK726 pKa = 9.05AHH728 pKa = 6.82PEE730 pKa = 4.04EE731 pKa = 4.65VVDD734 pKa = 4.61LALSRR739 pKa = 11.84LILIAVAGDD748 pKa = 3.4QLKK751 pKa = 10.75DD752 pKa = 3.46LNAIEE757 pKa = 5.11LIQYY761 pKa = 9.38GCADD765 pKa = 3.47VKK767 pKa = 11.17DD768 pKa = 3.9IFMKK772 pKa = 10.63PEE774 pKa = 3.47EE775 pKa = 4.68HH776 pKa = 6.58SPQKK780 pKa = 9.89IAEE783 pKa = 4.11GRR785 pKa = 11.84FRR787 pKa = 11.84LIWISSLIDD796 pKa = 3.4LTVQALLHH804 pKa = 6.2KK805 pKa = 10.06ADD807 pKa = 3.91NAAHH811 pKa = 5.94VDD813 pKa = 3.84AYY815 pKa = 10.79QAGHH819 pKa = 6.38LTCAALGMGHH829 pKa = 6.05SPEE832 pKa = 4.33GLQHH836 pKa = 6.79LVRR839 pKa = 11.84AFEE842 pKa = 4.23RR843 pKa = 11.84EE844 pKa = 4.08GVAEE848 pKa = 4.29RR849 pKa = 11.84NVSSDD854 pKa = 2.72ASAFDD859 pKa = 3.97LSIDD863 pKa = 3.47GSFIHH868 pKa = 7.52ADD870 pKa = 3.44GEE872 pKa = 4.27RR873 pKa = 11.84RR874 pKa = 11.84ADD876 pKa = 3.3NCADD880 pKa = 3.62EE881 pKa = 5.26DD882 pKa = 3.87VARR885 pKa = 11.84LVKK888 pKa = 10.27RR889 pKa = 11.84YY890 pKa = 9.69AHH892 pKa = 6.77ILCSHH897 pKa = 6.47VLNNHH902 pKa = 5.23GEE904 pKa = 3.85VWLCLKK910 pKa = 10.95YY911 pKa = 10.76GVTSSGQLSTTTQNTFSRR929 pKa = 11.84SVMAAYY935 pKa = 9.63GGCLGWTCAGDD946 pKa = 4.29DD947 pKa = 4.41LVGDD951 pKa = 4.22EE952 pKa = 5.03NFDD955 pKa = 3.93HH956 pKa = 7.04NRR958 pKa = 11.84LLHH961 pKa = 5.79FGVRR965 pKa = 11.84SRR967 pKa = 11.84DD968 pKa = 3.52VEE970 pKa = 4.29LHH972 pKa = 5.49VGEE975 pKa = 5.35ADD977 pKa = 3.83FTSHH981 pKa = 8.15LINTSTATAAFGNVEE996 pKa = 3.74KK997 pKa = 10.25MLWHH1001 pKa = 7.25LYY1003 pKa = 7.6DD1004 pKa = 3.16TCTDD1008 pKa = 2.79VSTNRR1013 pKa = 11.84EE1014 pKa = 3.88RR1015 pKa = 11.84FGSSLYY1021 pKa = 10.26ILRR1024 pKa = 11.84DD1025 pKa = 3.6TPGVLEE1031 pKa = 5.08DD1032 pKa = 3.52VTAITNDD1039 pKa = 3.27FGINTDD1045 pKa = 3.41GYY1047 pKa = 9.35VAEE1050 pKa = 4.62SSLIRR1055 pKa = 11.84DD1056 pKa = 4.03LAA1058 pKa = 5.32
MM1 pKa = 7.69ALLLSRR7 pKa = 11.84VQGADD12 pKa = 2.94TKK14 pKa = 10.84AVKK17 pKa = 10.29KK18 pKa = 10.57VVTSFGLEE26 pKa = 3.66PGAARR31 pKa = 11.84AYY33 pKa = 10.56LHH35 pKa = 6.84LGLLFGIYY43 pKa = 10.29KK44 pKa = 9.2LVCARR49 pKa = 11.84KK50 pKa = 8.91EE51 pKa = 3.91LCPPIARR58 pKa = 11.84ACLRR62 pKa = 11.84VLYY65 pKa = 9.55YY66 pKa = 9.36VVKK69 pKa = 9.48MSSPGVSLVSWLKK82 pKa = 9.44EE83 pKa = 3.39RR84 pKa = 11.84LRR86 pKa = 11.84RR87 pKa = 11.84GRR89 pKa = 11.84IADD92 pKa = 4.0CVQGGPPTKK101 pKa = 9.9HH102 pKa = 6.8GYY104 pKa = 9.14FRR106 pKa = 11.84WNGKK110 pKa = 9.96DD111 pKa = 3.63FVRR114 pKa = 11.84HH115 pKa = 5.34SDD117 pKa = 3.34EE118 pKa = 4.49SISRR122 pKa = 11.84VKK124 pKa = 9.93MLRR127 pKa = 11.84GTTTIDD133 pKa = 3.02AYY135 pKa = 10.38GQEE138 pKa = 4.02STAYY142 pKa = 8.56NWEE145 pKa = 3.82ILEE148 pKa = 4.06PARR151 pKa = 11.84PKK153 pKa = 10.67DD154 pKa = 3.65LSPGEE159 pKa = 4.19SALEE163 pKa = 4.05GLSSNARR170 pKa = 11.84PSPLEE175 pKa = 4.0KK176 pKa = 10.6GHH178 pKa = 6.18VLVHH182 pKa = 6.97DD183 pKa = 5.18SILNLFSTGLVLNNILIMSRR203 pKa = 11.84HH204 pKa = 4.61KK205 pKa = 9.45TASLTHH211 pKa = 5.78LVVRR215 pKa = 11.84GTNTTAVAGRR225 pKa = 11.84QSGVRR230 pKa = 11.84ISLDD234 pKa = 2.85RR235 pKa = 11.84AYY237 pKa = 11.35YY238 pKa = 10.69LDD240 pKa = 5.66DD241 pKa = 4.41FGAPKK246 pKa = 9.53WDD248 pKa = 4.14EE249 pKa = 4.0KK250 pKa = 11.21RR251 pKa = 11.84HH252 pKa = 5.02TCTFLDD258 pKa = 4.92FMAIPLDD265 pKa = 3.63KK266 pKa = 11.3DD267 pKa = 4.02EE268 pKa = 5.66ISMIGVKK275 pKa = 8.74SLKK278 pKa = 10.83DD279 pKa = 3.21KK280 pKa = 11.08DD281 pKa = 3.11ITRR284 pKa = 11.84NYY286 pKa = 8.76EE287 pKa = 3.93LEE289 pKa = 4.5RR290 pKa = 11.84GTITYY295 pKa = 8.1ATDD298 pKa = 3.42DD299 pKa = 3.73FGNVVKK305 pKa = 10.74EE306 pKa = 4.04EE307 pKa = 4.15GSIPEE312 pKa = 4.36AEE314 pKa = 4.07LHH316 pKa = 5.42IHH318 pKa = 6.3NLGLALAHH326 pKa = 6.73IVSQPGASSSGVGVVQNGYY345 pKa = 10.03KK346 pKa = 10.21LAGMWLGQPAHH357 pKa = 6.15SLKK360 pKa = 10.27KK361 pKa = 10.16HH362 pKa = 5.84RR363 pKa = 11.84GKK365 pKa = 10.97ANLFMHH371 pKa = 7.04TDD373 pKa = 3.69AIMANLEE380 pKa = 3.85QMGLYY385 pKa = 9.37KK386 pKa = 10.56HH387 pKa = 6.28PLIEE391 pKa = 6.31RR392 pKa = 11.84IQQWSDD398 pKa = 2.92SLQTPTSLPEE408 pKa = 3.91APGEE412 pKa = 4.07SRR414 pKa = 11.84EE415 pKa = 4.34SKK417 pKa = 10.43KK418 pKa = 10.46EE419 pKa = 3.32RR420 pKa = 11.84AARR423 pKa = 11.84KK424 pKa = 7.77WFEE427 pKa = 3.87YY428 pKa = 8.17YY429 pKa = 9.75QEE431 pKa = 4.58YY432 pKa = 10.6KK433 pKa = 10.75DD434 pKa = 3.9EE435 pKa = 4.89PYY437 pKa = 11.02EE438 pKa = 4.27AMEE441 pKa = 4.15EE442 pKa = 4.2DD443 pKa = 3.89EE444 pKa = 4.35EE445 pKa = 4.4EE446 pKa = 4.49EE447 pKa = 4.29YY448 pKa = 11.16EE449 pKa = 4.24KK450 pKa = 11.23VYY452 pKa = 10.72GRR454 pKa = 11.84SGHH457 pKa = 6.78ARR459 pKa = 11.84TQPAVALPKK468 pKa = 10.39AVAAPGPRR476 pKa = 11.84IPPRR480 pKa = 11.84ARR482 pKa = 11.84HH483 pKa = 5.84PGEE486 pKa = 4.12SAAEE490 pKa = 3.82RR491 pKa = 11.84ALEE494 pKa = 3.81ITRR497 pKa = 11.84QNMLLDD503 pKa = 3.34IDD505 pKa = 4.98RR506 pKa = 11.84NVKK509 pKa = 8.06SWKK512 pKa = 10.05YY513 pKa = 10.61GILHH517 pKa = 6.41EE518 pKa = 4.31EE519 pKa = 4.06APRR522 pKa = 11.84VLRR525 pKa = 11.84EE526 pKa = 3.25ISYY529 pKa = 10.23YY530 pKa = 10.32RR531 pKa = 11.84EE532 pKa = 3.58EE533 pKa = 3.82ATADD537 pKa = 3.06VRR539 pKa = 11.84EE540 pKa = 4.07RR541 pKa = 11.84LKK543 pKa = 11.14AVLATPSNALLRR555 pKa = 11.84DD556 pKa = 3.65YY557 pKa = 9.51MKK559 pKa = 10.36KK560 pKa = 10.21VSPMWSPGEE569 pKa = 3.92EE570 pKa = 4.2GEE572 pKa = 5.09EE573 pKa = 3.86ILDD576 pKa = 3.94RR577 pKa = 11.84DD578 pKa = 3.65GNPYY582 pKa = 9.25FRR584 pKa = 11.84RR585 pKa = 11.84VGSYY589 pKa = 10.63EE590 pKa = 3.62PTRR593 pKa = 11.84TAKK596 pKa = 9.76AKK598 pKa = 10.15KK599 pKa = 9.36YY600 pKa = 10.2RR601 pKa = 11.84EE602 pKa = 4.22EE603 pKa = 3.89EE604 pKa = 3.56TDD606 pKa = 3.25LQKK609 pKa = 11.0RR610 pKa = 11.84LRR612 pKa = 11.84EE613 pKa = 3.93LANRR617 pKa = 11.84YY618 pKa = 8.81YY619 pKa = 11.07ADD621 pKa = 4.24KK622 pKa = 11.28LHH624 pKa = 6.21GCKK627 pKa = 9.88KK628 pKa = 10.0GEE630 pKa = 3.82YY631 pKa = 9.57RR632 pKa = 11.84IPTSSKK638 pKa = 10.48KK639 pKa = 10.85NIDD642 pKa = 3.6RR643 pKa = 11.84SLKK646 pKa = 9.86AQAALAVAGPPQLTSEE662 pKa = 4.1QEE664 pKa = 4.23EE665 pKa = 4.32EE666 pKa = 4.04FEE668 pKa = 4.02AAVRR672 pKa = 11.84LVRR675 pKa = 11.84EE676 pKa = 4.32KK677 pKa = 10.87YY678 pKa = 9.57KK679 pKa = 11.02AGIGDD684 pKa = 4.19EE685 pKa = 4.45PIKK688 pKa = 10.2TYY690 pKa = 10.92LEE692 pKa = 4.13EE693 pKa = 5.78GDD695 pKa = 3.83YY696 pKa = 11.64GFLKK700 pKa = 10.14TFLGFEE706 pKa = 4.53DD707 pKa = 4.05KK708 pKa = 11.14SSGVSARR715 pKa = 11.84FRR717 pKa = 11.84NMKK720 pKa = 8.88KK721 pKa = 9.9AAWVKK726 pKa = 9.05AHH728 pKa = 6.82PEE730 pKa = 4.04EE731 pKa = 4.65VVDD734 pKa = 4.61LALSRR739 pKa = 11.84LILIAVAGDD748 pKa = 3.4QLKK751 pKa = 10.75DD752 pKa = 3.46LNAIEE757 pKa = 5.11LIQYY761 pKa = 9.38GCADD765 pKa = 3.47VKK767 pKa = 11.17DD768 pKa = 3.9IFMKK772 pKa = 10.63PEE774 pKa = 3.47EE775 pKa = 4.68HH776 pKa = 6.58SPQKK780 pKa = 9.89IAEE783 pKa = 4.11GRR785 pKa = 11.84FRR787 pKa = 11.84LIWISSLIDD796 pKa = 3.4LTVQALLHH804 pKa = 6.2KK805 pKa = 10.06ADD807 pKa = 3.91NAAHH811 pKa = 5.94VDD813 pKa = 3.84AYY815 pKa = 10.79QAGHH819 pKa = 6.38LTCAALGMGHH829 pKa = 6.05SPEE832 pKa = 4.33GLQHH836 pKa = 6.79LVRR839 pKa = 11.84AFEE842 pKa = 4.23RR843 pKa = 11.84EE844 pKa = 4.08GVAEE848 pKa = 4.29RR849 pKa = 11.84NVSSDD854 pKa = 2.72ASAFDD859 pKa = 3.97LSIDD863 pKa = 3.47GSFIHH868 pKa = 7.52ADD870 pKa = 3.44GEE872 pKa = 4.27RR873 pKa = 11.84RR874 pKa = 11.84ADD876 pKa = 3.3NCADD880 pKa = 3.62EE881 pKa = 5.26DD882 pKa = 3.87VARR885 pKa = 11.84LVKK888 pKa = 10.27RR889 pKa = 11.84YY890 pKa = 9.69AHH892 pKa = 6.77ILCSHH897 pKa = 6.47VLNNHH902 pKa = 5.23GEE904 pKa = 3.85VWLCLKK910 pKa = 10.95YY911 pKa = 10.76GVTSSGQLSTTTQNTFSRR929 pKa = 11.84SVMAAYY935 pKa = 9.63GGCLGWTCAGDD946 pKa = 4.29DD947 pKa = 4.41LVGDD951 pKa = 4.22EE952 pKa = 5.03NFDD955 pKa = 3.93HH956 pKa = 7.04NRR958 pKa = 11.84LLHH961 pKa = 5.79FGVRR965 pKa = 11.84SRR967 pKa = 11.84DD968 pKa = 3.52VEE970 pKa = 4.29LHH972 pKa = 5.49VGEE975 pKa = 5.35ADD977 pKa = 3.83FTSHH981 pKa = 8.15LINTSTATAAFGNVEE996 pKa = 3.74KK997 pKa = 10.25MLWHH1001 pKa = 7.25LYY1003 pKa = 7.6DD1004 pKa = 3.16TCTDD1008 pKa = 2.79VSTNRR1013 pKa = 11.84EE1014 pKa = 3.88RR1015 pKa = 11.84FGSSLYY1021 pKa = 10.26ILRR1024 pKa = 11.84DD1025 pKa = 3.6TPGVLEE1031 pKa = 5.08DD1032 pKa = 3.52VTAITNDD1039 pKa = 3.27FGINTDD1045 pKa = 3.41GYY1047 pKa = 9.35VAEE1050 pKa = 4.62SSLIRR1055 pKa = 11.84DD1056 pKa = 4.03LAA1058 pKa = 5.32
Molecular weight: 118.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEA0|A0A1L3KEA0_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 5 OX=1922702 PE=4 SV=1
MM1 pKa = 7.3VSPGKK6 pKa = 9.88RR7 pKa = 11.84VKK9 pKa = 9.85RR10 pKa = 11.84TIRR13 pKa = 11.84KK14 pKa = 8.73QKK16 pKa = 10.44AKK18 pKa = 10.34VIKK21 pKa = 10.38DD22 pKa = 3.31VTLLRR27 pKa = 11.84GIKK30 pKa = 9.65QGVGMVVKK38 pKa = 10.52RR39 pKa = 11.84PFGSAKK45 pKa = 9.9ARR47 pKa = 11.84PRR49 pKa = 11.84RR50 pKa = 11.84TANRR54 pKa = 11.84GRR56 pKa = 11.84NISKK60 pKa = 10.48KK61 pKa = 10.33AALCALTNAHH71 pKa = 6.62LPLPRR76 pKa = 11.84AVGAYY81 pKa = 8.15TVVKK85 pKa = 7.22TTTVIDD91 pKa = 3.75SASRR95 pKa = 11.84AMLFGTIKK103 pKa = 10.62GNRR106 pKa = 11.84SNHH109 pKa = 6.9PDD111 pKa = 3.57PAWYY115 pKa = 8.21DD116 pKa = 3.95TVAVTAGTSSTTAVGAADD134 pKa = 3.51NAKK137 pKa = 10.27FYY139 pKa = 11.07LDD141 pKa = 3.44TALASSGFASCRR153 pKa = 11.84LVPAAFTVQVMCPKK167 pKa = 10.39SLQTADD173 pKa = 3.76GIVYY177 pKa = 10.28IGRR180 pKa = 11.84CKK182 pKa = 10.12QVLDD186 pKa = 3.9LMGNTRR192 pKa = 11.84TWTTVMDD199 pKa = 4.62DD200 pKa = 3.66LVSYY204 pKa = 10.31SSPRR208 pKa = 11.84LCSAGKK214 pKa = 9.54LALRR218 pKa = 11.84GVKK221 pKa = 9.8VDD223 pKa = 5.31AIPNNMSQLSDD234 pKa = 3.67FCPRR238 pKa = 11.84AILTSSDD245 pKa = 3.34TNKK248 pKa = 8.09WTWGQGGVDD257 pKa = 3.32AQFEE261 pKa = 4.47GFAPIFVYY269 pKa = 10.67NKK271 pKa = 8.71NTVDD275 pKa = 3.58LQYY278 pKa = 11.29LVTVEE283 pKa = 3.4WRR285 pKa = 11.84VRR287 pKa = 11.84FDD289 pKa = 3.68PGNPAYY295 pKa = 10.13AGHH298 pKa = 6.56TFHH301 pKa = 7.23PVSSDD306 pKa = 3.62TCWSEE311 pKa = 3.83TLKK314 pKa = 11.07GMEE317 pKa = 4.18AEE319 pKa = 4.18GHH321 pKa = 5.7GVVDD325 pKa = 4.46LAEE328 pKa = 5.24GIADD332 pKa = 4.04FGDD335 pKa = 3.58AAMTAVGALLL345 pKa = 3.8
MM1 pKa = 7.3VSPGKK6 pKa = 9.88RR7 pKa = 11.84VKK9 pKa = 9.85RR10 pKa = 11.84TIRR13 pKa = 11.84KK14 pKa = 8.73QKK16 pKa = 10.44AKK18 pKa = 10.34VIKK21 pKa = 10.38DD22 pKa = 3.31VTLLRR27 pKa = 11.84GIKK30 pKa = 9.65QGVGMVVKK38 pKa = 10.52RR39 pKa = 11.84PFGSAKK45 pKa = 9.9ARR47 pKa = 11.84PRR49 pKa = 11.84RR50 pKa = 11.84TANRR54 pKa = 11.84GRR56 pKa = 11.84NISKK60 pKa = 10.48KK61 pKa = 10.33AALCALTNAHH71 pKa = 6.62LPLPRR76 pKa = 11.84AVGAYY81 pKa = 8.15TVVKK85 pKa = 7.22TTTVIDD91 pKa = 3.75SASRR95 pKa = 11.84AMLFGTIKK103 pKa = 10.62GNRR106 pKa = 11.84SNHH109 pKa = 6.9PDD111 pKa = 3.57PAWYY115 pKa = 8.21DD116 pKa = 3.95TVAVTAGTSSTTAVGAADD134 pKa = 3.51NAKK137 pKa = 10.27FYY139 pKa = 11.07LDD141 pKa = 3.44TALASSGFASCRR153 pKa = 11.84LVPAAFTVQVMCPKK167 pKa = 10.39SLQTADD173 pKa = 3.76GIVYY177 pKa = 10.28IGRR180 pKa = 11.84CKK182 pKa = 10.12QVLDD186 pKa = 3.9LMGNTRR192 pKa = 11.84TWTTVMDD199 pKa = 4.62DD200 pKa = 3.66LVSYY204 pKa = 10.31SSPRR208 pKa = 11.84LCSAGKK214 pKa = 9.54LALRR218 pKa = 11.84GVKK221 pKa = 9.8VDD223 pKa = 5.31AIPNNMSQLSDD234 pKa = 3.67FCPRR238 pKa = 11.84AILTSSDD245 pKa = 3.34TNKK248 pKa = 8.09WTWGQGGVDD257 pKa = 3.32AQFEE261 pKa = 4.47GFAPIFVYY269 pKa = 10.67NKK271 pKa = 8.71NTVDD275 pKa = 3.58LQYY278 pKa = 11.29LVTVEE283 pKa = 3.4WRR285 pKa = 11.84VRR287 pKa = 11.84FDD289 pKa = 3.68PGNPAYY295 pKa = 10.13AGHH298 pKa = 6.56TFHH301 pKa = 7.23PVSSDD306 pKa = 3.62TCWSEE311 pKa = 3.83TLKK314 pKa = 11.07GMEE317 pKa = 4.18AEE319 pKa = 4.18GHH321 pKa = 5.7GVVDD325 pKa = 4.46LAEE328 pKa = 5.24GIADD332 pKa = 4.04FGDD335 pKa = 3.58AAMTAVGALLL345 pKa = 3.8
Molecular weight: 37.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1403 |
345 |
1058 |
701.5 |
77.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.979 ± 0.846 | 1.497 ± 0.279 |
5.845 ± 0.025 | 6.415 ± 2.447 |
2.922 ± 0.291 | 7.627 ± 0.408 |
2.851 ± 0.734 | 4.277 ± 0.418 |
5.916 ± 0.09 | 9.337 ± 1.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.996 ± 0.321 | 3.35 ± 0.219 |
4.205 ± 0.226 | 2.495 ± 0.06 |
6.771 ± 0.358 | 6.842 ± 0.092 |
5.845 ± 1.492 | 6.985 ± 1.351 |
1.426 ± 0.164 | 3.421 ± 0.577 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
