
Methanocorpusculum labreanum (strain ATCC 43576 / DSM 4855 / Z)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanocorpusculaceae; Methanocorpusculum; Methanocorpusculum labreanum
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
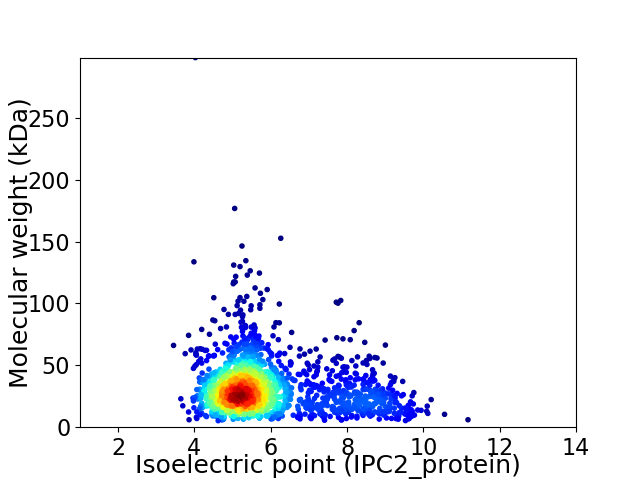
Virtual 2D-PAGE plot for 1739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A2STJ3|RNP4_METLZ Ribonuclease P protein component 4 OS=Methanocorpusculum labreanum (strain ATCC 43576 / DSM 4855 / Z) OX=410358 GN=rnp4 PE=3 SV=1
MM1 pKa = 7.6ISNGVVQKK9 pKa = 9.24TLILGGILLLLITAMIVPVSASSEE33 pKa = 4.2NYY35 pKa = 9.96LVFYY39 pKa = 10.18MVGSDD44 pKa = 3.7LEE46 pKa = 4.38SGEE49 pKa = 4.47DD50 pKa = 3.51HH51 pKa = 6.64LASMNLNDD59 pKa = 5.33LVNNLDD65 pKa = 4.05PEE67 pKa = 4.18ITDD70 pKa = 3.17ILVIYY75 pKa = 10.23GGADD79 pKa = 3.0QTGWDD84 pKa = 3.75NGLSITNLDD93 pKa = 4.16LLKK96 pKa = 10.51KK97 pKa = 10.14DD98 pKa = 4.15LEE100 pKa = 4.65DD101 pKa = 5.18GIVGSDD107 pKa = 4.3EE108 pKa = 5.34DD109 pKa = 4.07PQTPTEE115 pKa = 4.07YY116 pKa = 11.09VLIRR120 pKa = 11.84ISDD123 pKa = 3.67VDD125 pKa = 3.35ISSPGALSEE134 pKa = 3.99GLQYY138 pKa = 11.24AEE140 pKa = 5.5SYY142 pKa = 10.81SADD145 pKa = 3.51NDD147 pKa = 3.75LASANRR153 pKa = 11.84YY154 pKa = 9.31LLFWNHH160 pKa = 5.2GGGYY164 pKa = 9.93EE165 pKa = 4.19GFGANEE171 pKa = 4.77LFDD174 pKa = 3.39TWMSVDD180 pKa = 4.27DD181 pKa = 4.66LQTGLAASKK190 pKa = 9.69PYY192 pKa = 10.5DD193 pKa = 3.57IIIFDD198 pKa = 4.41ACQMASLEE206 pKa = 4.31VADD209 pKa = 5.59ALDD212 pKa = 3.46THH214 pKa = 7.83ADD216 pKa = 3.41FLLASEE222 pKa = 4.77EE223 pKa = 4.07NVPGIGLNYY232 pKa = 10.1VGIGKK237 pKa = 9.67ALSSDD242 pKa = 3.21PAMTPEE248 pKa = 4.17EE249 pKa = 4.15VGKK252 pKa = 10.67SIISAYY258 pKa = 10.65VSGTNEE264 pKa = 3.55NRR266 pKa = 11.84KK267 pKa = 7.2TLSLVRR273 pKa = 11.84LQKK276 pKa = 10.7AKK278 pKa = 10.94NVVASLDD285 pKa = 3.62VLGTSLKK292 pKa = 10.87DD293 pKa = 3.31SLNNEE298 pKa = 4.28DD299 pKa = 4.58SLAALGDD306 pKa = 3.42IYY308 pKa = 9.55TTVQGYY314 pKa = 9.25GRR316 pKa = 11.84SDD318 pKa = 3.44DD319 pKa = 3.81GSRR322 pKa = 11.84VASVDD327 pKa = 3.71LYY329 pKa = 11.4QFADD333 pKa = 4.8LIYY336 pKa = 10.88ANTEE340 pKa = 3.85DD341 pKa = 3.62TTEE344 pKa = 3.92LHH346 pKa = 6.3RR347 pKa = 11.84AAGSLMTSLQDD358 pKa = 3.45YY359 pKa = 9.7VVAADD364 pKa = 3.84HH365 pKa = 6.08STHH368 pKa = 6.53FSAANGVSIAAPKK381 pKa = 10.62EE382 pKa = 4.14DD383 pKa = 3.64FTKK386 pKa = 10.78AIPQSIAFGRR396 pKa = 11.84NGWYY400 pKa = 9.9EE401 pKa = 3.93YY402 pKa = 10.45FSSYY406 pKa = 8.56MSKK409 pKa = 9.02TGTQSQPSASYY420 pKa = 10.9KK421 pKa = 10.66SGNTDD426 pKa = 2.18SRR428 pKa = 11.84TIVVHH433 pKa = 7.72DD434 pKa = 3.96DD435 pKa = 4.08TEE437 pKa = 4.72TTWAFADD444 pKa = 3.54YY445 pKa = 10.94VYY447 pKa = 10.13YY448 pKa = 10.42TDD450 pKa = 3.23SNYY453 pKa = 10.0IIVGQTPLEE462 pKa = 4.37EE463 pKa = 4.64IYY465 pKa = 10.96APSEE469 pKa = 4.19DD470 pKa = 4.18GLWISVPTGEE480 pKa = 4.81YY481 pKa = 10.56EE482 pKa = 4.01EE483 pKa = 5.2PDD485 pKa = 2.95WDD487 pKa = 3.99GSWFVLRR494 pKa = 11.84SDD496 pKa = 4.32GKK498 pKa = 9.86NADD501 pKa = 3.68VLVSMTYY508 pKa = 10.1AYY510 pKa = 10.57SVVEE514 pKa = 4.36DD515 pKa = 4.06EE516 pKa = 4.65SVKK519 pKa = 10.66DD520 pKa = 3.43VYY522 pKa = 10.95LIYY525 pKa = 11.05GNITRR530 pKa = 11.84EE531 pKa = 4.08INGKK535 pKa = 10.05DD536 pKa = 2.81ITHH539 pKa = 7.18PSVITAVVDD548 pKa = 3.55MNAMKK553 pKa = 10.19TNYY556 pKa = 10.06LFVEE560 pKa = 4.8SVSEE564 pKa = 4.23DD565 pKa = 2.66GWYY568 pKa = 10.69SRR570 pKa = 11.84TNLWGDD576 pKa = 3.65TKK578 pKa = 11.1LLPGDD583 pKa = 3.69VFTPLLEE590 pKa = 4.35VYY592 pKa = 10.47NEE594 pKa = 4.11EE595 pKa = 4.31EE596 pKa = 4.11EE597 pKa = 4.73SISTVYY603 pKa = 10.73SADD606 pKa = 3.22SFTFGDD612 pKa = 5.04DD613 pKa = 3.39PAEE616 pKa = 4.23DD617 pKa = 3.91LVFTALDD624 pKa = 3.63EE625 pKa = 4.64NNCYY629 pKa = 9.41WVVEE633 pKa = 4.06LDD635 pKa = 5.59DD636 pKa = 5.05YY637 pKa = 11.91LDD639 pKa = 5.17DD640 pKa = 4.41DD641 pKa = 4.25AFYY644 pKa = 11.16LEE646 pKa = 5.42EE647 pKa = 5.42PDD649 pKa = 5.44FPDD652 pKa = 3.08TGAAKK657 pKa = 10.5SPILPAGMFFGLAAAFLLIKK677 pKa = 10.44RR678 pKa = 11.84RR679 pKa = 3.68
MM1 pKa = 7.6ISNGVVQKK9 pKa = 9.24TLILGGILLLLITAMIVPVSASSEE33 pKa = 4.2NYY35 pKa = 9.96LVFYY39 pKa = 10.18MVGSDD44 pKa = 3.7LEE46 pKa = 4.38SGEE49 pKa = 4.47DD50 pKa = 3.51HH51 pKa = 6.64LASMNLNDD59 pKa = 5.33LVNNLDD65 pKa = 4.05PEE67 pKa = 4.18ITDD70 pKa = 3.17ILVIYY75 pKa = 10.23GGADD79 pKa = 3.0QTGWDD84 pKa = 3.75NGLSITNLDD93 pKa = 4.16LLKK96 pKa = 10.51KK97 pKa = 10.14DD98 pKa = 4.15LEE100 pKa = 4.65DD101 pKa = 5.18GIVGSDD107 pKa = 4.3EE108 pKa = 5.34DD109 pKa = 4.07PQTPTEE115 pKa = 4.07YY116 pKa = 11.09VLIRR120 pKa = 11.84ISDD123 pKa = 3.67VDD125 pKa = 3.35ISSPGALSEE134 pKa = 3.99GLQYY138 pKa = 11.24AEE140 pKa = 5.5SYY142 pKa = 10.81SADD145 pKa = 3.51NDD147 pKa = 3.75LASANRR153 pKa = 11.84YY154 pKa = 9.31LLFWNHH160 pKa = 5.2GGGYY164 pKa = 9.93EE165 pKa = 4.19GFGANEE171 pKa = 4.77LFDD174 pKa = 3.39TWMSVDD180 pKa = 4.27DD181 pKa = 4.66LQTGLAASKK190 pKa = 9.69PYY192 pKa = 10.5DD193 pKa = 3.57IIIFDD198 pKa = 4.41ACQMASLEE206 pKa = 4.31VADD209 pKa = 5.59ALDD212 pKa = 3.46THH214 pKa = 7.83ADD216 pKa = 3.41FLLASEE222 pKa = 4.77EE223 pKa = 4.07NVPGIGLNYY232 pKa = 10.1VGIGKK237 pKa = 9.67ALSSDD242 pKa = 3.21PAMTPEE248 pKa = 4.17EE249 pKa = 4.15VGKK252 pKa = 10.67SIISAYY258 pKa = 10.65VSGTNEE264 pKa = 3.55NRR266 pKa = 11.84KK267 pKa = 7.2TLSLVRR273 pKa = 11.84LQKK276 pKa = 10.7AKK278 pKa = 10.94NVVASLDD285 pKa = 3.62VLGTSLKK292 pKa = 10.87DD293 pKa = 3.31SLNNEE298 pKa = 4.28DD299 pKa = 4.58SLAALGDD306 pKa = 3.42IYY308 pKa = 9.55TTVQGYY314 pKa = 9.25GRR316 pKa = 11.84SDD318 pKa = 3.44DD319 pKa = 3.81GSRR322 pKa = 11.84VASVDD327 pKa = 3.71LYY329 pKa = 11.4QFADD333 pKa = 4.8LIYY336 pKa = 10.88ANTEE340 pKa = 3.85DD341 pKa = 3.62TTEE344 pKa = 3.92LHH346 pKa = 6.3RR347 pKa = 11.84AAGSLMTSLQDD358 pKa = 3.45YY359 pKa = 9.7VVAADD364 pKa = 3.84HH365 pKa = 6.08STHH368 pKa = 6.53FSAANGVSIAAPKK381 pKa = 10.62EE382 pKa = 4.14DD383 pKa = 3.64FTKK386 pKa = 10.78AIPQSIAFGRR396 pKa = 11.84NGWYY400 pKa = 9.9EE401 pKa = 3.93YY402 pKa = 10.45FSSYY406 pKa = 8.56MSKK409 pKa = 9.02TGTQSQPSASYY420 pKa = 10.9KK421 pKa = 10.66SGNTDD426 pKa = 2.18SRR428 pKa = 11.84TIVVHH433 pKa = 7.72DD434 pKa = 3.96DD435 pKa = 4.08TEE437 pKa = 4.72TTWAFADD444 pKa = 3.54YY445 pKa = 10.94VYY447 pKa = 10.13YY448 pKa = 10.42TDD450 pKa = 3.23SNYY453 pKa = 10.0IIVGQTPLEE462 pKa = 4.37EE463 pKa = 4.64IYY465 pKa = 10.96APSEE469 pKa = 4.19DD470 pKa = 4.18GLWISVPTGEE480 pKa = 4.81YY481 pKa = 10.56EE482 pKa = 4.01EE483 pKa = 5.2PDD485 pKa = 2.95WDD487 pKa = 3.99GSWFVLRR494 pKa = 11.84SDD496 pKa = 4.32GKK498 pKa = 9.86NADD501 pKa = 3.68VLVSMTYY508 pKa = 10.1AYY510 pKa = 10.57SVVEE514 pKa = 4.36DD515 pKa = 4.06EE516 pKa = 4.65SVKK519 pKa = 10.66DD520 pKa = 3.43VYY522 pKa = 10.95LIYY525 pKa = 11.05GNITRR530 pKa = 11.84EE531 pKa = 4.08INGKK535 pKa = 10.05DD536 pKa = 2.81ITHH539 pKa = 7.18PSVITAVVDD548 pKa = 3.55MNAMKK553 pKa = 10.19TNYY556 pKa = 10.06LFVEE560 pKa = 4.8SVSEE564 pKa = 4.23DD565 pKa = 2.66GWYY568 pKa = 10.69SRR570 pKa = 11.84TNLWGDD576 pKa = 3.65TKK578 pKa = 11.1LLPGDD583 pKa = 3.69VFTPLLEE590 pKa = 4.35VYY592 pKa = 10.47NEE594 pKa = 4.11EE595 pKa = 4.31EE596 pKa = 4.11EE597 pKa = 4.73SISTVYY603 pKa = 10.73SADD606 pKa = 3.22SFTFGDD612 pKa = 5.04DD613 pKa = 3.39PAEE616 pKa = 4.23DD617 pKa = 3.91LVFTALDD624 pKa = 3.63EE625 pKa = 4.64NNCYY629 pKa = 9.41WVVEE633 pKa = 4.06LDD635 pKa = 5.59DD636 pKa = 5.05YY637 pKa = 11.91LDD639 pKa = 5.17DD640 pKa = 4.41DD641 pKa = 4.25AFYY644 pKa = 11.16LEE646 pKa = 5.42EE647 pKa = 5.42PDD649 pKa = 5.44FPDD652 pKa = 3.08TGAAKK657 pKa = 10.5SPILPAGMFFGLAAAFLLIKK677 pKa = 10.44RR678 pKa = 11.84RR679 pKa = 3.68
Molecular weight: 74.24 kDa
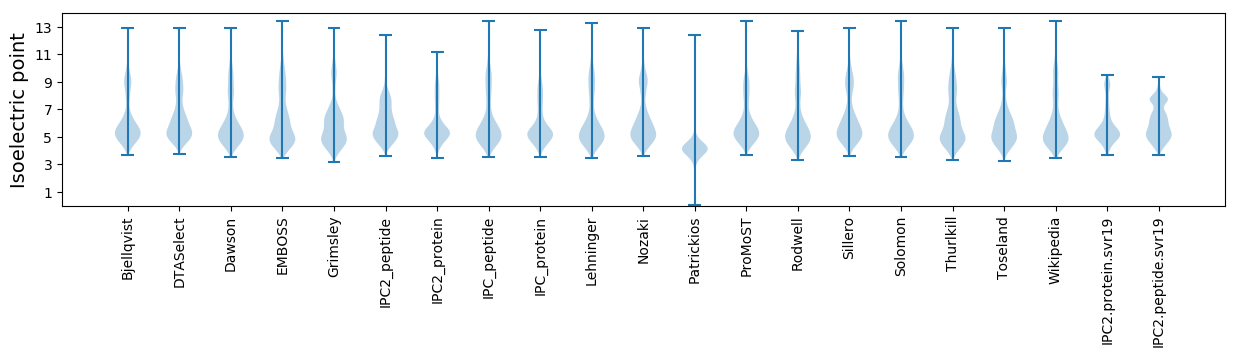
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A2STJ4|A2STJ4_METLZ 5'-phosphoribosylglycinamide transformylase OS=Methanocorpusculum labreanum (strain ATCC 43576 / DSM 4855 / Z) OX=410358 GN=Mlab_1486 PE=3 SV=1
MM1 pKa = 7.66SKK3 pKa = 8.88LTKK6 pKa = 9.77SRR8 pKa = 11.84KK9 pKa = 8.87IRR11 pKa = 11.84LAKK14 pKa = 9.4ATQQNRR20 pKa = 11.84RR21 pKa = 11.84VPAWVMIKK29 pKa = 9.0TKK31 pKa = 9.53RR32 pKa = 11.84TVVSHH37 pKa = 5.98PKK39 pKa = 8.67RR40 pKa = 11.84RR41 pKa = 11.84NWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.56VV50 pKa = 3.07
MM1 pKa = 7.66SKK3 pKa = 8.88LTKK6 pKa = 9.77SRR8 pKa = 11.84KK9 pKa = 8.87IRR11 pKa = 11.84LAKK14 pKa = 9.4ATQQNRR20 pKa = 11.84RR21 pKa = 11.84VPAWVMIKK29 pKa = 9.0TKK31 pKa = 9.53RR32 pKa = 11.84TVVSHH37 pKa = 5.98PKK39 pKa = 8.67RR40 pKa = 11.84RR41 pKa = 11.84NWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.56VV50 pKa = 3.07
Molecular weight: 6.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
522529 |
47 |
2772 |
300.5 |
33.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.525 ± 0.064 | 1.507 ± 0.027 |
5.526 ± 0.049 | 6.7 ± 0.055 |
3.93 ± 0.044 | 7.769 ± 0.052 |
1.835 ± 0.025 | 7.631 ± 0.057 |
5.504 ± 0.051 | 9.202 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.865 ± 0.029 | 3.562 ± 0.043 |
4.434 ± 0.032 | 2.512 ± 0.027 |
4.678 ± 0.051 | 6.03 ± 0.043 |
5.977 ± 0.065 | 7.476 ± 0.055 |
0.923 ± 0.021 | 3.416 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
