
Christensenella minuta
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Christensenellaceae; Christensenella
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
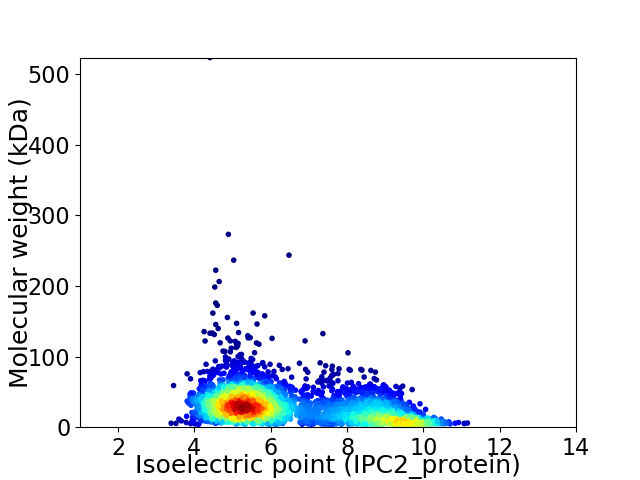
Virtual 2D-PAGE plot for 3119 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A136Q5C9|A0A136Q5C9_9FIRM RND transporter HAE1/HME family permease protein OS=Christensenella minuta OX=626937 GN=HMPREF3293_01478 PE=4 SV=1
MM1 pKa = 7.9DD2 pKa = 3.76RR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 10.34RR6 pKa = 11.84FLTMLVCFVLAAFMLTACAGGGAPAATEE34 pKa = 4.23SVAAGQGAEE43 pKa = 4.34TEE45 pKa = 4.57TPAATDD51 pKa = 3.23TAEE54 pKa = 4.65AGTEE58 pKa = 3.96SSPAILTSADD68 pKa = 3.36VGDD71 pKa = 3.76VSGIEE76 pKa = 4.19GWDD79 pKa = 3.74GPVEE83 pKa = 4.34SQVMTAGEE91 pKa = 4.8DD92 pKa = 3.56GDD94 pKa = 3.91QFGTAEE100 pKa = 4.12STVVVGKK107 pKa = 10.35GPKK110 pKa = 10.35GEE112 pKa = 4.56VSTPWTEE119 pKa = 3.47IALTDD124 pKa = 3.71EE125 pKa = 4.65EE126 pKa = 4.81KK127 pKa = 11.01EE128 pKa = 4.11QIRR131 pKa = 11.84SGNYY135 pKa = 7.99KK136 pKa = 10.26AAICFHH142 pKa = 6.16YY143 pKa = 8.22TTDD146 pKa = 2.97DD147 pKa = 3.57WYY149 pKa = 10.78VMQRR153 pKa = 11.84KK154 pKa = 9.31GIEE157 pKa = 3.81EE158 pKa = 4.15TFADD162 pKa = 4.01LGIEE166 pKa = 4.34VVAVTDD172 pKa = 3.76AGFSAEE178 pKa = 4.03QQVSDD183 pKa = 3.99IEE185 pKa = 4.57NVLEE189 pKa = 4.45LNPDD193 pKa = 4.07IIISIPVDD201 pKa = 3.47EE202 pKa = 4.94TSTAVAYY209 pKa = 10.11QKK211 pKa = 11.12AADD214 pKa = 3.96AGVKK218 pKa = 9.87LVFMDD223 pKa = 3.98NCPSNMTAGKK233 pKa = 10.13DD234 pKa = 3.61YY235 pKa = 11.23VSIVAADD242 pKa = 4.1RR243 pKa = 11.84YY244 pKa = 9.0GCGMICADD252 pKa = 4.73LMARR256 pKa = 11.84EE257 pKa = 4.37LNYY260 pKa = 10.45EE261 pKa = 3.96GTVGVIYY268 pKa = 10.68YY269 pKa = 10.32DD270 pKa = 3.34ADD272 pKa = 3.74FFVTTRR278 pKa = 11.84DD279 pKa = 3.17CAGFVDD285 pKa = 4.96RR286 pKa = 11.84MKK288 pKa = 10.94LKK290 pKa = 10.39YY291 pKa = 9.95PNIEE295 pKa = 3.83IVTEE299 pKa = 3.96MGFADD304 pKa = 3.52ATLSGEE310 pKa = 4.34VADD313 pKa = 5.9AMLAQYY319 pKa = 10.49PDD321 pKa = 3.07IDD323 pKa = 4.86GIFGSWDD330 pKa = 3.15VPAEE334 pKa = 4.16YY335 pKa = 9.88IASSASAVGRR345 pKa = 11.84DD346 pKa = 3.7DD347 pKa = 5.82LVVNCVGLGTTTASMTAGDD366 pKa = 4.8GIINTISTQRR376 pKa = 11.84PYY378 pKa = 11.4DD379 pKa = 3.52QGCTEE384 pKa = 5.1ALLACYY390 pKa = 10.26GLLGKK395 pKa = 9.7DD396 pKa = 2.91APEE399 pKa = 4.25YY400 pKa = 9.32VTVPALGVTKK410 pKa = 10.44EE411 pKa = 4.02GSEE414 pKa = 4.06NSASLKK420 pKa = 10.38DD421 pKa = 3.85AYY423 pKa = 11.06QLFYY427 pKa = 11.02QIDD430 pKa = 4.05TLPDD434 pKa = 3.0WLQQLMM440 pKa = 4.09
MM1 pKa = 7.9DD2 pKa = 3.76RR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 10.34RR6 pKa = 11.84FLTMLVCFVLAAFMLTACAGGGAPAATEE34 pKa = 4.23SVAAGQGAEE43 pKa = 4.34TEE45 pKa = 4.57TPAATDD51 pKa = 3.23TAEE54 pKa = 4.65AGTEE58 pKa = 3.96SSPAILTSADD68 pKa = 3.36VGDD71 pKa = 3.76VSGIEE76 pKa = 4.19GWDD79 pKa = 3.74GPVEE83 pKa = 4.34SQVMTAGEE91 pKa = 4.8DD92 pKa = 3.56GDD94 pKa = 3.91QFGTAEE100 pKa = 4.12STVVVGKK107 pKa = 10.35GPKK110 pKa = 10.35GEE112 pKa = 4.56VSTPWTEE119 pKa = 3.47IALTDD124 pKa = 3.71EE125 pKa = 4.65EE126 pKa = 4.81KK127 pKa = 11.01EE128 pKa = 4.11QIRR131 pKa = 11.84SGNYY135 pKa = 7.99KK136 pKa = 10.26AAICFHH142 pKa = 6.16YY143 pKa = 8.22TTDD146 pKa = 2.97DD147 pKa = 3.57WYY149 pKa = 10.78VMQRR153 pKa = 11.84KK154 pKa = 9.31GIEE157 pKa = 3.81EE158 pKa = 4.15TFADD162 pKa = 4.01LGIEE166 pKa = 4.34VVAVTDD172 pKa = 3.76AGFSAEE178 pKa = 4.03QQVSDD183 pKa = 3.99IEE185 pKa = 4.57NVLEE189 pKa = 4.45LNPDD193 pKa = 4.07IIISIPVDD201 pKa = 3.47EE202 pKa = 4.94TSTAVAYY209 pKa = 10.11QKK211 pKa = 11.12AADD214 pKa = 3.96AGVKK218 pKa = 9.87LVFMDD223 pKa = 3.98NCPSNMTAGKK233 pKa = 10.13DD234 pKa = 3.61YY235 pKa = 11.23VSIVAADD242 pKa = 4.1RR243 pKa = 11.84YY244 pKa = 9.0GCGMICADD252 pKa = 4.73LMARR256 pKa = 11.84EE257 pKa = 4.37LNYY260 pKa = 10.45EE261 pKa = 3.96GTVGVIYY268 pKa = 10.68YY269 pKa = 10.32DD270 pKa = 3.34ADD272 pKa = 3.74FFVTTRR278 pKa = 11.84DD279 pKa = 3.17CAGFVDD285 pKa = 4.96RR286 pKa = 11.84MKK288 pKa = 10.94LKK290 pKa = 10.39YY291 pKa = 9.95PNIEE295 pKa = 3.83IVTEE299 pKa = 3.96MGFADD304 pKa = 3.52ATLSGEE310 pKa = 4.34VADD313 pKa = 5.9AMLAQYY319 pKa = 10.49PDD321 pKa = 3.07IDD323 pKa = 4.86GIFGSWDD330 pKa = 3.15VPAEE334 pKa = 4.16YY335 pKa = 9.88IASSASAVGRR345 pKa = 11.84DD346 pKa = 3.7DD347 pKa = 5.82LVVNCVGLGTTTASMTAGDD366 pKa = 4.8GIINTISTQRR376 pKa = 11.84PYY378 pKa = 11.4DD379 pKa = 3.52QGCTEE384 pKa = 5.1ALLACYY390 pKa = 10.26GLLGKK395 pKa = 9.7DD396 pKa = 2.91APEE399 pKa = 4.25YY400 pKa = 9.32VTVPALGVTKK410 pKa = 10.44EE411 pKa = 4.02GSEE414 pKa = 4.06NSASLKK420 pKa = 10.38DD421 pKa = 3.85AYY423 pKa = 11.06QLFYY427 pKa = 11.02QIDD430 pKa = 4.05TLPDD434 pKa = 3.0WLQQLMM440 pKa = 4.09
Molecular weight: 46.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A136Q814|A0A136Q814_9FIRM DEAD-box ATP-dependent RNA helicase CshA family protein OS=Christensenella minuta OX=626937 GN=HMPREF3293_00367 PE=3 SV=1
MM1 pKa = 7.31KK2 pKa = 10.07RR3 pKa = 11.84KK4 pKa = 9.67KK5 pKa = 10.65SPWRR9 pKa = 11.84MLHH12 pKa = 5.97GRR14 pKa = 11.84GISNVKK20 pKa = 9.99HH21 pKa = 5.61IRR23 pKa = 11.84KK24 pKa = 9.58APGTLSGAFPRR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84GTGQRR44 pKa = 11.84LLTVAFFF51 pKa = 4.35
MM1 pKa = 7.31KK2 pKa = 10.07RR3 pKa = 11.84KK4 pKa = 9.67KK5 pKa = 10.65SPWRR9 pKa = 11.84MLHH12 pKa = 5.97GRR14 pKa = 11.84GISNVKK20 pKa = 9.99HH21 pKa = 5.61IRR23 pKa = 11.84KK24 pKa = 9.58APGTLSGAFPRR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84GTGQRR44 pKa = 11.84LLTVAFFF51 pKa = 4.35
Molecular weight: 5.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
890553 |
29 |
4909 |
285.5 |
31.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.169 ± 0.053 | 1.466 ± 0.021 |
5.478 ± 0.035 | 7.202 ± 0.049 |
4.079 ± 0.036 | 8.03 ± 0.042 |
1.628 ± 0.02 | 6.821 ± 0.045 |
6.143 ± 0.042 | 8.803 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.101 ± 0.027 | 3.807 ± 0.026 |
3.75 ± 0.028 | 3.136 ± 0.023 |
5.069 ± 0.044 | 5.406 ± 0.037 |
5.347 ± 0.051 | 7.093 ± 0.039 |
0.838 ± 0.014 | 3.636 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
