
Human papillomavirus type 41
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Nupapillomavirus; Nupapillomavirus 1
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
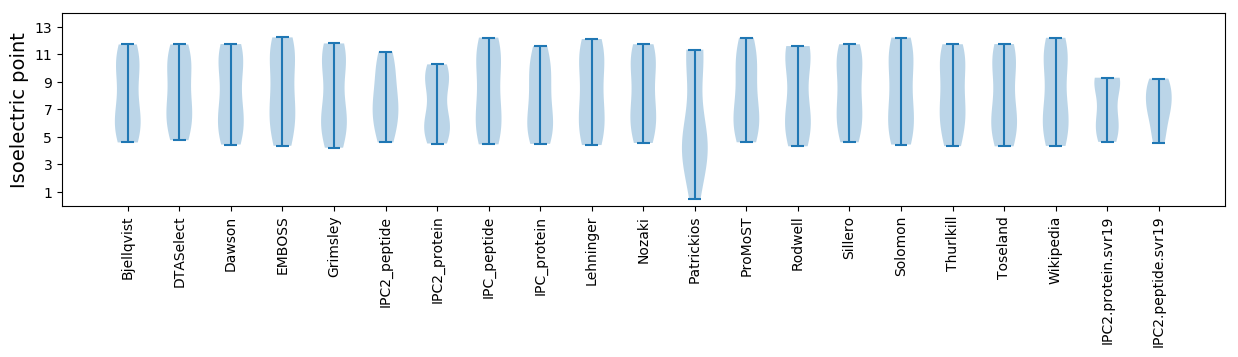
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q84212|Q84212_HPV41 ORF X protein OS=Human papillomavirus type 41 OX=10589 GN=ORF X PE=4 SV=1
MM1 pKa = 7.91LARR4 pKa = 11.84QRR6 pKa = 11.84VKK8 pKa = 10.55RR9 pKa = 11.84ANPEE13 pKa = 3.72QLYY16 pKa = 7.9KK17 pKa = 9.76TCKK20 pKa = 9.13ATGGDD25 pKa = 4.25CPPDD29 pKa = 3.39VIKK32 pKa = 10.42RR33 pKa = 11.84YY34 pKa = 8.47EE35 pKa = 3.94QTTPADD41 pKa = 4.02SILKK45 pKa = 8.54YY46 pKa = 10.62GSVGVFFGGLGIGTGRR62 pKa = 11.84GGGGTVLGAGAVGGRR77 pKa = 11.84PSISSGAIGPRR88 pKa = 11.84DD89 pKa = 3.54ILPIEE94 pKa = 4.46SGGPSLAEE102 pKa = 4.58EE103 pKa = 4.38IPLLPMAPRR112 pKa = 11.84VPRR115 pKa = 11.84PTDD118 pKa = 3.24PFRR121 pKa = 11.84PSVLEE126 pKa = 3.72EE127 pKa = 3.88PFIIRR132 pKa = 11.84PPEE135 pKa = 4.03RR136 pKa = 11.84PNILHH141 pKa = 6.06EE142 pKa = 4.25QRR144 pKa = 11.84FPTDD148 pKa = 3.15AAPFDD153 pKa = 3.97NGNTEE158 pKa = 3.59ITTIPSQYY166 pKa = 9.93DD167 pKa = 3.38VSGGGVDD174 pKa = 3.21IQIIEE179 pKa = 4.4LPSVNDD185 pKa = 4.13PGPSVVTRR193 pKa = 11.84TQYY196 pKa = 11.34NNPTFEE202 pKa = 4.56VEE204 pKa = 4.0VSTDD208 pKa = 2.85ISGEE212 pKa = 4.05TSSTDD217 pKa = 3.49NIIVGAEE224 pKa = 3.86SGGTSVGDD232 pKa = 3.56NAEE235 pKa = 5.23LIPLLDD241 pKa = 3.53ISRR244 pKa = 11.84GDD246 pKa = 3.89TIDD249 pKa = 3.45TTILAPGEE257 pKa = 4.16EE258 pKa = 4.11EE259 pKa = 4.2TAFVTSTPEE268 pKa = 3.37RR269 pKa = 11.84VPIQEE274 pKa = 4.26RR275 pKa = 11.84LPIRR279 pKa = 11.84PYY281 pKa = 10.18GRR283 pKa = 11.84QYY285 pKa = 9.82QQVRR289 pKa = 11.84VTDD292 pKa = 4.05PEE294 pKa = 4.22FLDD297 pKa = 3.78SAAVLVSLEE306 pKa = 4.14NPVFDD311 pKa = 5.69ADD313 pKa = 3.3ITLTFEE319 pKa = 6.62DD320 pKa = 5.33DD321 pKa = 3.35LQQALRR327 pKa = 11.84SDD329 pKa = 3.33TDD331 pKa = 3.38LRR333 pKa = 11.84DD334 pKa = 3.33VRR336 pKa = 11.84RR337 pKa = 11.84LSRR340 pKa = 11.84PYY342 pKa = 8.82YY343 pKa = 9.08QRR345 pKa = 11.84RR346 pKa = 11.84TTGLRR351 pKa = 11.84VSRR354 pKa = 11.84LGQRR358 pKa = 11.84RR359 pKa = 11.84GTISTRR365 pKa = 11.84SGVQVGSAAHH375 pKa = 6.6FFQDD379 pKa = 4.14ISPIGQAIEE388 pKa = 4.66PIDD391 pKa = 5.0AIEE394 pKa = 4.52LDD396 pKa = 3.81VLGEE400 pKa = 3.94QSGEE404 pKa = 3.98GTIVRR409 pKa = 11.84GDD411 pKa = 3.61PTPSIEE417 pKa = 4.35QDD419 pKa = 2.75IGLTALGDD427 pKa = 3.94NIEE430 pKa = 4.67NEE432 pKa = 4.11LQEE435 pKa = 5.33IDD437 pKa = 5.17LLTADD442 pKa = 4.51GEE444 pKa = 4.29EE445 pKa = 4.23DD446 pKa = 3.53QEE448 pKa = 4.88GRR450 pKa = 11.84DD451 pKa = 3.69LQLVFSTGNDD461 pKa = 3.09EE462 pKa = 4.65VVDD465 pKa = 3.51IMTIPIRR472 pKa = 11.84AGGDD476 pKa = 3.22DD477 pKa = 3.88RR478 pKa = 11.84PSVFIFSDD486 pKa = 3.72DD487 pKa = 3.49GTHH490 pKa = 6.18IVYY493 pKa = 7.71PTSTTATTPLVPAQPSDD510 pKa = 3.43VPYY513 pKa = 10.47IVVDD517 pKa = 4.9LYY519 pKa = 11.18SGSMDD524 pKa = 3.9YY525 pKa = 10.7DD526 pKa = 3.35IHH528 pKa = 6.94PSLLRR533 pKa = 11.84RR534 pKa = 11.84KK535 pKa = 8.4RR536 pKa = 11.84KK537 pKa = 8.73KK538 pKa = 9.56RR539 pKa = 11.84KK540 pKa = 8.97RR541 pKa = 11.84VYY543 pKa = 10.53FSDD546 pKa = 3.09GRR548 pKa = 11.84VASRR552 pKa = 11.84PKK554 pKa = 10.56
MM1 pKa = 7.91LARR4 pKa = 11.84QRR6 pKa = 11.84VKK8 pKa = 10.55RR9 pKa = 11.84ANPEE13 pKa = 3.72QLYY16 pKa = 7.9KK17 pKa = 9.76TCKK20 pKa = 9.13ATGGDD25 pKa = 4.25CPPDD29 pKa = 3.39VIKK32 pKa = 10.42RR33 pKa = 11.84YY34 pKa = 8.47EE35 pKa = 3.94QTTPADD41 pKa = 4.02SILKK45 pKa = 8.54YY46 pKa = 10.62GSVGVFFGGLGIGTGRR62 pKa = 11.84GGGGTVLGAGAVGGRR77 pKa = 11.84PSISSGAIGPRR88 pKa = 11.84DD89 pKa = 3.54ILPIEE94 pKa = 4.46SGGPSLAEE102 pKa = 4.58EE103 pKa = 4.38IPLLPMAPRR112 pKa = 11.84VPRR115 pKa = 11.84PTDD118 pKa = 3.24PFRR121 pKa = 11.84PSVLEE126 pKa = 3.72EE127 pKa = 3.88PFIIRR132 pKa = 11.84PPEE135 pKa = 4.03RR136 pKa = 11.84PNILHH141 pKa = 6.06EE142 pKa = 4.25QRR144 pKa = 11.84FPTDD148 pKa = 3.15AAPFDD153 pKa = 3.97NGNTEE158 pKa = 3.59ITTIPSQYY166 pKa = 9.93DD167 pKa = 3.38VSGGGVDD174 pKa = 3.21IQIIEE179 pKa = 4.4LPSVNDD185 pKa = 4.13PGPSVVTRR193 pKa = 11.84TQYY196 pKa = 11.34NNPTFEE202 pKa = 4.56VEE204 pKa = 4.0VSTDD208 pKa = 2.85ISGEE212 pKa = 4.05TSSTDD217 pKa = 3.49NIIVGAEE224 pKa = 3.86SGGTSVGDD232 pKa = 3.56NAEE235 pKa = 5.23LIPLLDD241 pKa = 3.53ISRR244 pKa = 11.84GDD246 pKa = 3.89TIDD249 pKa = 3.45TTILAPGEE257 pKa = 4.16EE258 pKa = 4.11EE259 pKa = 4.2TAFVTSTPEE268 pKa = 3.37RR269 pKa = 11.84VPIQEE274 pKa = 4.26RR275 pKa = 11.84LPIRR279 pKa = 11.84PYY281 pKa = 10.18GRR283 pKa = 11.84QYY285 pKa = 9.82QQVRR289 pKa = 11.84VTDD292 pKa = 4.05PEE294 pKa = 4.22FLDD297 pKa = 3.78SAAVLVSLEE306 pKa = 4.14NPVFDD311 pKa = 5.69ADD313 pKa = 3.3ITLTFEE319 pKa = 6.62DD320 pKa = 5.33DD321 pKa = 3.35LQQALRR327 pKa = 11.84SDD329 pKa = 3.33TDD331 pKa = 3.38LRR333 pKa = 11.84DD334 pKa = 3.33VRR336 pKa = 11.84RR337 pKa = 11.84LSRR340 pKa = 11.84PYY342 pKa = 8.82YY343 pKa = 9.08QRR345 pKa = 11.84RR346 pKa = 11.84TTGLRR351 pKa = 11.84VSRR354 pKa = 11.84LGQRR358 pKa = 11.84RR359 pKa = 11.84GTISTRR365 pKa = 11.84SGVQVGSAAHH375 pKa = 6.6FFQDD379 pKa = 4.14ISPIGQAIEE388 pKa = 4.66PIDD391 pKa = 5.0AIEE394 pKa = 4.52LDD396 pKa = 3.81VLGEE400 pKa = 3.94QSGEE404 pKa = 3.98GTIVRR409 pKa = 11.84GDD411 pKa = 3.61PTPSIEE417 pKa = 4.35QDD419 pKa = 2.75IGLTALGDD427 pKa = 3.94NIEE430 pKa = 4.67NEE432 pKa = 4.11LQEE435 pKa = 5.33IDD437 pKa = 5.17LLTADD442 pKa = 4.51GEE444 pKa = 4.29EE445 pKa = 4.23DD446 pKa = 3.53QEE448 pKa = 4.88GRR450 pKa = 11.84DD451 pKa = 3.69LQLVFSTGNDD461 pKa = 3.09EE462 pKa = 4.65VVDD465 pKa = 3.51IMTIPIRR472 pKa = 11.84AGGDD476 pKa = 3.22DD477 pKa = 3.88RR478 pKa = 11.84PSVFIFSDD486 pKa = 3.72DD487 pKa = 3.49GTHH490 pKa = 6.18IVYY493 pKa = 7.71PTSTTATTPLVPAQPSDD510 pKa = 3.43VPYY513 pKa = 10.47IVVDD517 pKa = 4.9LYY519 pKa = 11.18SGSMDD524 pKa = 3.9YY525 pKa = 10.7DD526 pKa = 3.35IHH528 pKa = 6.94PSLLRR533 pKa = 11.84RR534 pKa = 11.84KK535 pKa = 8.4RR536 pKa = 11.84KK537 pKa = 8.73KK538 pKa = 9.56RR539 pKa = 11.84KK540 pKa = 8.97RR541 pKa = 11.84VYY543 pKa = 10.53FSDD546 pKa = 3.09GRR548 pKa = 11.84VASRR552 pKa = 11.84PKK554 pKa = 10.56
Molecular weight: 59.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84214|Q84214_HPV41 ORF Z protein OS=Human papillomavirus type 41 OX=10589 GN=ORF Z PE=4 SV=1
MM1 pKa = 7.64LPLTVCLLLDD11 pKa = 3.19IHH13 pKa = 6.02FTILLMRR20 pKa = 11.84MAKK23 pKa = 9.38RR24 pKa = 11.84WSLKK28 pKa = 10.27FPLISSGPSVSVSQIPIPLHH48 pKa = 5.63FVISPFLTLTRR59 pKa = 11.84SVWSGVFVGLRR70 pKa = 11.84FLGDD74 pKa = 3.59SPP76 pKa = 4.73
MM1 pKa = 7.64LPLTVCLLLDD11 pKa = 3.19IHH13 pKa = 6.02FTILLMRR20 pKa = 11.84MAKK23 pKa = 9.38RR24 pKa = 11.84WSLKK28 pKa = 10.27FPLISSGPSVSVSQIPIPLHH48 pKa = 5.63FVISPFLTLTRR59 pKa = 11.84SVWSGVFVGLRR70 pKa = 11.84FLGDD74 pKa = 3.59SPP76 pKa = 4.73
Molecular weight: 8.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2765 |
72 |
614 |
251.4 |
28.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.004 ± 0.263 | 2.061 ± 0.64 |
6.004 ± 0.821 | 6.221 ± 0.683 |
4.014 ± 0.619 | 6.727 ± 0.868 |
2.242 ± 0.474 | 5.461 ± 0.677 |
4.34 ± 0.678 | 9.042 ± 0.739 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.17 ± 0.537 | 4.087 ± 0.701 |
5.497 ± 0.834 | 4.304 ± 0.429 |
7.74 ± 0.789 | 7.414 ± 0.338 |
6.293 ± 0.574 | 5.931 ± 0.523 |
1.193 ± 0.338 | 3.255 ± 0.372 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
