
Actinomadura sp. RB29
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Actinomadura
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
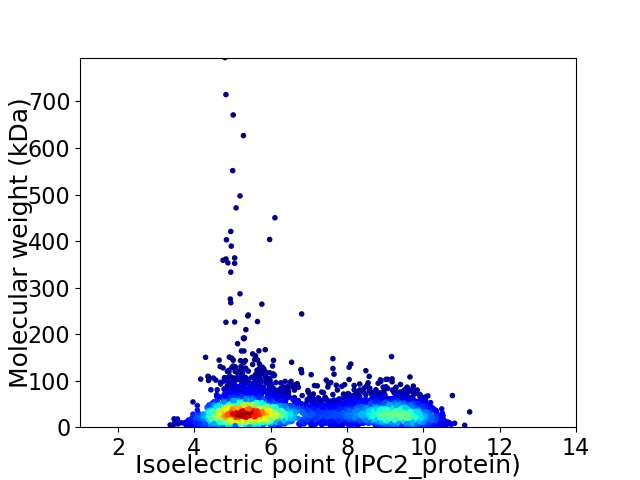
Virtual 2D-PAGE plot for 5700 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
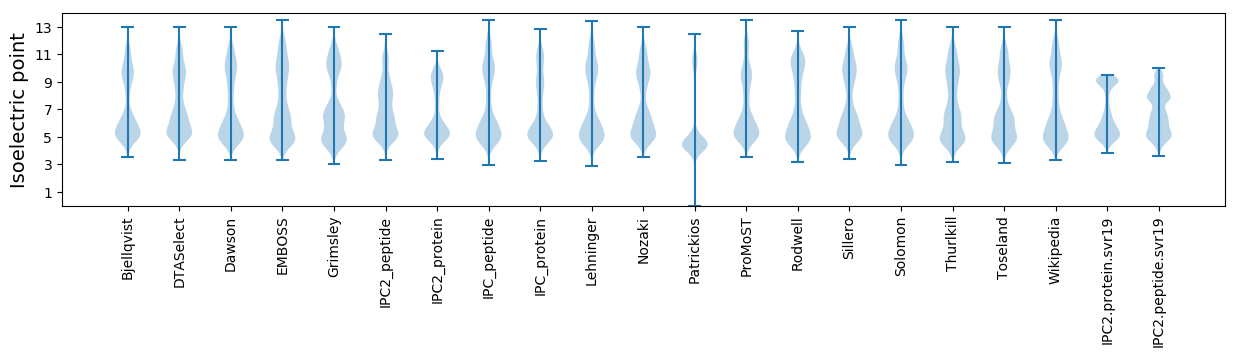
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P4UD21|A0A2P4UD21_9ACTN Ferrous-iron efflux pump FieF OS=Actinomadura sp. RB29 OX=1926885 GN=fieF_2 PE=4 SV=1
MM1 pKa = 7.37SPSTILRR8 pKa = 11.84QAAADD13 pKa = 3.52AEE15 pKa = 4.47FRR17 pKa = 11.84AEE19 pKa = 4.33LLDD22 pKa = 3.79NPEE25 pKa = 4.14VFGVAAGTVPEE36 pKa = 4.27SVEE39 pKa = 4.17RR40 pKa = 11.84QDD42 pKa = 4.36EE43 pKa = 4.08ASLNYY48 pKa = 7.48WTEE51 pKa = 3.59GVAAVDD57 pKa = 5.03AYY59 pKa = 10.66ACSSTCTSGPFTFACDD75 pKa = 3.54GTTKK79 pKa = 10.8GG80 pKa = 3.96
MM1 pKa = 7.37SPSTILRR8 pKa = 11.84QAAADD13 pKa = 3.52AEE15 pKa = 4.47FRR17 pKa = 11.84AEE19 pKa = 4.33LLDD22 pKa = 3.79NPEE25 pKa = 4.14VFGVAAGTVPEE36 pKa = 4.27SVEE39 pKa = 4.17RR40 pKa = 11.84QDD42 pKa = 4.36EE43 pKa = 4.08ASLNYY48 pKa = 7.48WTEE51 pKa = 3.59GVAAVDD57 pKa = 5.03AYY59 pKa = 10.66ACSSTCTSGPFTFACDD75 pKa = 3.54GTTKK79 pKa = 10.8GG80 pKa = 3.96
Molecular weight: 8.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P4UEC8|A0A2P4UEC8_9ACTN Alpha/beta hydrolase family protein OS=Actinomadura sp. RB29 OX=1926885 GN=BTM25_45700 PE=4 SV=1
MM1 pKa = 7.44RR2 pKa = 11.84RR3 pKa = 11.84ARR5 pKa = 11.84HH6 pKa = 5.58LRR8 pKa = 11.84RR9 pKa = 11.84ITDD12 pKa = 3.71TARR15 pKa = 11.84TACPRR20 pKa = 11.84RR21 pKa = 11.84AARR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84AVRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84TRR35 pKa = 11.84GPRR38 pKa = 11.84SIPGLRR44 pKa = 11.84GTPSRR49 pKa = 11.84RR50 pKa = 11.84SKK52 pKa = 10.44LRR54 pKa = 11.84PRR56 pKa = 11.84RR57 pKa = 11.84VPRR60 pKa = 11.84LLRR63 pKa = 11.84TPRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84AAGRR72 pKa = 11.84LRR74 pKa = 11.84RR75 pKa = 11.84AVGVCPRR82 pKa = 11.84ARR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84PLVLCLTLSLSPRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84QAPRR105 pKa = 11.84PRR107 pKa = 11.84RR108 pKa = 11.84VRR110 pKa = 11.84RR111 pKa = 11.84LPRR114 pKa = 11.84TRR116 pKa = 11.84LPRR119 pKa = 11.84TRR121 pKa = 11.84RR122 pKa = 11.84TGAVGRR128 pKa = 11.84PRR130 pKa = 11.84RR131 pKa = 11.84VARR134 pKa = 11.84ARR136 pKa = 11.84RR137 pKa = 11.84VLRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84LLVVRR148 pKa = 11.84LTLVLRR154 pKa = 11.84ARR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84PSGAARR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84PSPRR171 pKa = 11.84TRR173 pKa = 11.84PARR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84MPHH181 pKa = 6.07PAGLRR186 pKa = 11.84ALRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84TTRR195 pKa = 11.84TCRR198 pKa = 11.84RR199 pKa = 11.84AGLCRR204 pKa = 11.84TRR206 pKa = 11.84RR207 pKa = 11.84VRR209 pKa = 11.84RR210 pKa = 11.84TVSRR214 pKa = 11.84RR215 pKa = 11.84LRR217 pKa = 11.84MRR219 pKa = 11.84AGARR223 pKa = 11.84GTGSIAGRR231 pKa = 11.84NGGGLAAGGSAPGRR245 pKa = 11.84RR246 pKa = 11.84CGARR250 pKa = 11.84PRR252 pKa = 11.84LRR254 pKa = 11.84PRR256 pKa = 11.84PRR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84VGLRR264 pKa = 11.84PRR266 pKa = 11.84LRR268 pKa = 11.84LRR270 pKa = 11.84VDD272 pKa = 3.14LRR274 pKa = 11.84LRR276 pKa = 11.84LGACPP281 pKa = 3.84
MM1 pKa = 7.44RR2 pKa = 11.84RR3 pKa = 11.84ARR5 pKa = 11.84HH6 pKa = 5.58LRR8 pKa = 11.84RR9 pKa = 11.84ITDD12 pKa = 3.71TARR15 pKa = 11.84TACPRR20 pKa = 11.84RR21 pKa = 11.84AARR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84AVRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84TRR35 pKa = 11.84GPRR38 pKa = 11.84SIPGLRR44 pKa = 11.84GTPSRR49 pKa = 11.84RR50 pKa = 11.84SKK52 pKa = 10.44LRR54 pKa = 11.84PRR56 pKa = 11.84RR57 pKa = 11.84VPRR60 pKa = 11.84LLRR63 pKa = 11.84TPRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84AAGRR72 pKa = 11.84LRR74 pKa = 11.84RR75 pKa = 11.84AVGVCPRR82 pKa = 11.84ARR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84PLVLCLTLSLSPRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84QAPRR105 pKa = 11.84PRR107 pKa = 11.84RR108 pKa = 11.84VRR110 pKa = 11.84RR111 pKa = 11.84LPRR114 pKa = 11.84TRR116 pKa = 11.84LPRR119 pKa = 11.84TRR121 pKa = 11.84RR122 pKa = 11.84TGAVGRR128 pKa = 11.84PRR130 pKa = 11.84RR131 pKa = 11.84VARR134 pKa = 11.84ARR136 pKa = 11.84RR137 pKa = 11.84VLRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84LLVVRR148 pKa = 11.84LTLVLRR154 pKa = 11.84ARR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84PSGAARR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84PSPRR171 pKa = 11.84TRR173 pKa = 11.84PARR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84MPHH181 pKa = 6.07PAGLRR186 pKa = 11.84ALRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84TTRR195 pKa = 11.84TCRR198 pKa = 11.84RR199 pKa = 11.84AGLCRR204 pKa = 11.84TRR206 pKa = 11.84RR207 pKa = 11.84VRR209 pKa = 11.84RR210 pKa = 11.84TVSRR214 pKa = 11.84RR215 pKa = 11.84LRR217 pKa = 11.84MRR219 pKa = 11.84AGARR223 pKa = 11.84GTGSIAGRR231 pKa = 11.84NGGGLAAGGSAPGRR245 pKa = 11.84RR246 pKa = 11.84CGARR250 pKa = 11.84PRR252 pKa = 11.84LRR254 pKa = 11.84PRR256 pKa = 11.84PRR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84VGLRR264 pKa = 11.84PRR266 pKa = 11.84LRR268 pKa = 11.84LRR270 pKa = 11.84VDD272 pKa = 3.14LRR274 pKa = 11.84LRR276 pKa = 11.84LGACPP281 pKa = 3.84
Molecular weight: 32.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1924993 |
29 |
7640 |
337.7 |
36.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.652 ± 0.055 | 0.73 ± 0.01 |
6.322 ± 0.027 | 5.227 ± 0.033 |
2.791 ± 0.017 | 9.489 ± 0.036 |
2.161 ± 0.016 | 3.094 ± 0.021 |
1.829 ± 0.025 | 10.517 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.608 ± 0.015 | 1.637 ± 0.016 |
6.47 ± 0.036 | 2.293 ± 0.018 |
8.819 ± 0.044 | 4.448 ± 0.024 |
5.79 ± 0.03 | 8.772 ± 0.033 |
1.424 ± 0.013 | 1.927 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
